DNA and RNA sequencing has become the new watchword for the biological researchers, as this tells them exactly what is going on at cellular level in health and disease. However, it is a partial revelation unless we also know what is happening at the level of proteins as well. This could soon be happening, according to a new study published in the journal Nature Biotechnology on December 16, 2019. The researchers used nanopores to identify many of the amino acids that make up all the natural proteins in the world. This is a big advance towards being able to find the sequence in which they are found within proteins.
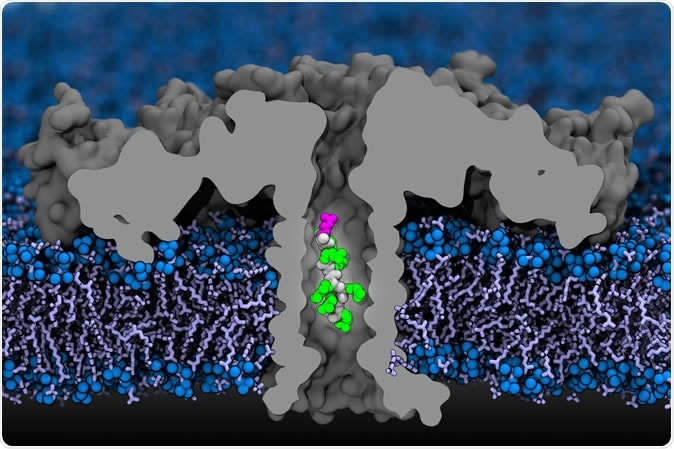
In this computer simulation, a portion of a protein moves through an aerolysin nanopore. The researchers used detailed simulations that mapped each atom, and confirmed their findings experimentally. Credit: Aleksei Aksimentiev
Knowing the differences between DNA sequencing and protein sequencing is crucial to understand how the body works. The DNA and its transcribed copy, the RNA, contain the encoded blueprint for every protein that can be produced in the organism, whether or not they are actually produced or not. These nucleic acids are therefore reflections of the potential of each cell to synthesize proteins.
On the other hand, the protein products created by the genes are the real workhorses that do the actual work of the cell. These proteins are created in accordance with the basic pattern or sequence of amino acid-coding bases laid down in the DNA, but they are far more than just a chain of amino acids. They are intensively modified after the fundamental work of splicing together the right sequence of amino acids is done. This chain is then chemically altered, cut and spliced, folded in several complex patterns, and so on. This is how the actual final protein product is fashioned.
The DNA molecule is a template for making copies of itself, and this process is therefore comparatively straightforward. On the other hand, proteins are designed to be created by the cell machinery in a one-by-one process. This makes it difficult to churn out copies of proteins as well as to read them.
There are 20 amino acids in proteins, compared to only 4 bases in DNA. Each of the 20 is also capable of undergoing alterations to become slightly different, while the protein is being produced and folded. Amino acids like leucine and isoleucine are also more difficult to differentiate in that their molecular weight and their atomic composition are identical, but the atoms are positioned differently.
Nanopore sequencing
To sequence DNA, nanopores are widely used. Nanopores are tiny protein channels forming part of a membrane. It was thought that amino acids differed too slightly from each other to be identified using nanopores, but the present study shows that they can.
As a nanopore, the researchers used a membrane channel called aerolysin, naturally occurring in bacteria. They did a two-part experiment using both computer modeling and actual laboratory-based experiments. In the latter, they ground protein very fine and then applied a chemical carrier to force the amino acids in the protein bits into the nanopore or channel.
The simulations run on this situation showed the presence of an inherent pocket that fits just one molecule in the aerolysin. This traps a single amino acid bound to the carrier within the nanopore’s sensing region.
The advantage of using the carrier molecule is that it keeps the amino acids long enough in the nanopore that the slight but distinctive electrical potentials, or ionic currents, produced by each amino acid, to be registered.
Aerolysin nanopore
The findings and follow-up
The results were breathtaking: the device rendered such sensitive measurements that it could identify 13/20 amino acids in nature. It showed the potential to identify hundreds of modified amino acids in its present form. However, says study leader Aleksei Aksimentiev, different forms of the nanopore can be used to increase the breadth of detection still further.
The researchers also modified the measurement tool to increase the sensitivity of detection of modified amino acids. They also tested the effects of chemically treating the protein first to make it easier to distinguish between different amino acids. This could help them identify all 20 amino acids in future.
Implications and future directions
The current work is described as only “a proof-of-concept study showing that we can identify the different amino acids.”
In contrast to the currently used method called mass spectrometry, which relies on comparing the sample being analyzed to the existing library of samples, to find a match, but lacks the power to actually determine the sequence, the use of nanopores is able to detect new forms or variants of the protein, and thus identify new mutations in the encoding genes. Says Aksimentiev, “With nanopores, we finally could look at those modifications which have not yet been studied.”
Researcher Abdelghani Oukhaled describes the effect of the research: “This work builds confidence and reassures the nanopore community that protein sequencing is indeed possible.”
The benefit of using aerolysin as the nanopore is that it can easily be fitted into existing nanopore installations, making it almost ready-to-use by other scientists. The next challenge for the researchers is to see if they can sequence the protein by identifying the amino acids in stepwise fashion, as each of them is isolated from the protein in order of occurrence.
Other applications are also being mulled over, such as using a combination of nanopores with immunoassays to pick out only those proteins that are being studied, and sequence these selectively. The aim is to understand if these proteins have been mutated or are intact, helping with clinical diagnosis.
The researchers are indeed excited at the increasingly minute accuracy with which they can now define biological molecules. They say, “Very likely, one day we will be able to tell the molecular makeup of the cell – what we are made of, down to the level of individual atoms.”
Journal reference:
Ouldali, H., Sarthak, K., Ensslen, T. et al. Electrical recognition of the twenty proteinogenic amino acids using an aerolysin nanopore. Nat Biotechnol (2019) doi:10.1038/s41587-019-0345-2, https://www.nature.com/articles/s41587-019-0345-2