The folding state of the proteins in live cells often reflect the cell’s general health. Australian scientists have developed a molecular probe that senses the state of the proteome—the entire set of the proteins—by measuring the polarity of the protein environment. The fluorescence signal of the probe quantifies unfolding and its chameleon-like color shift maps the cellular regions of enhanced misfolding, says the study published in the journal Angewandte Chemie.
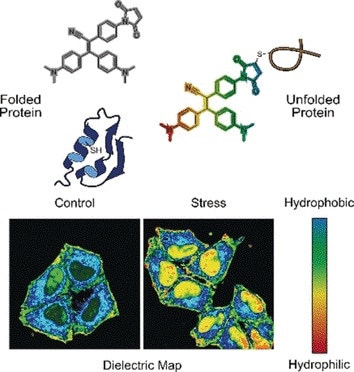
If live cells are stressed, protein-synthesis and folding-correction mechanisms are out of balance. Misfolded proteins remain stuck, enhanced degradation occurs, and inactive proteins and protein debris aggregate to form granules and condensates in the cytoplasm. Such aggregates play an important role in neurodegenerative diseases and cancer. One driving factor for the aggregation of misfolded proteins seems to be the polarity—the electronic distribution in an environment. Yuning Hong and colleagues at La Trobe University Melbourne and The University of Melbourne, Australia, have designed a two-modal fluorogenic probe to monitor protein aggregation in greater detail.
In one mode, the probe senses misfolded proteins. Correctly folded proteins are often stabilized by bridges made of the amino acid cysteine. These bridges are usually deeply buried, whereas misfolded proteins expose the cysteine residues at the surface. When the probe binds to cysteine exposed by a misfolded protein chain, fluorescence is switched on, explain the authors.
In the other mode, the probe assesses the polarity. Polar environments indicate an unbalanced electronic distribution, which can be measured by the dielectric constant. To measure this parameter, the researchers added an electronic “push–pull” chemical group to the fluorogenic probe. They observed that, in polar solutions with a high dielectric constant, the fluorogenic probe called NTPAN-MI emitted its fluorescence signal with a color shift. This “chameleon-like” color change thus indicates a polarity change.
The authors tested the NTPAN-MI probe on a human cell line, which they stressed by adding drugs that interfered with protein synthesis and folding. The scientists observed normal fluorescence in untreated cells, but bright fluorescence when unfolded or misfolded proteins accumulated in cells treated with toxins or infected by virus. In addition, the color shift signaled the polarity of the environment and thus the proteome state of each cellular compartment. The researchers reported that they visualized the “unfolded protein load” in the nucleus for the first time. Previous methods could only measure unfolded proteins in the cytoplasm.
With its two sensing modes—measuring unfolding and the polarity of the protein environment—the NTPAN-MI probe provides a sharper picture of the stress responses of live cells than what can be obtained with only one-modal probes or different methods. The authors point out that their method would allow scientists to obtain more accurate knowledge of the crosstalk of the cellular components in response to stress.
Source:
Journal reference:
Owyong, T. C., et al. (2019) A Molecular Chameleon for Mapping Subcellular Polarity in an Unfolded Proteome Environment. Angewandte Chemie International Edition. doi.org/10.1002/anie.201914263.