SARS-CoV-2 is responsible for the COVID-19 pandemic, which has sickened over 3.18 million people and taken more than 227,000 lives the world over. The novel coronavirus is a member of the same family from which the causative agents of the SARS and MERS outbreaks came.
.jpg)
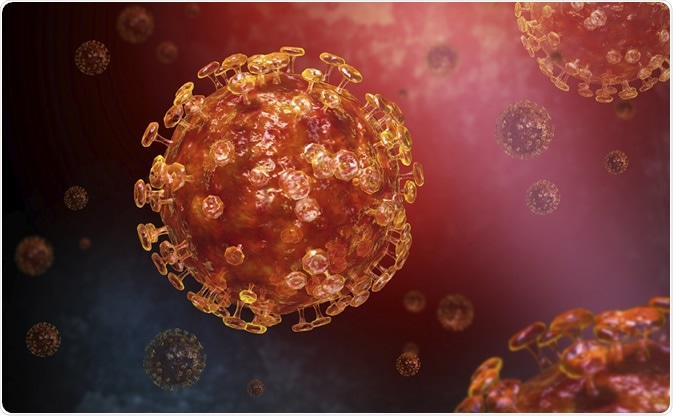
Novel Coronavirus SARS-CoV-2 Transmission electron micrograph of a SARS-CoV-2 virus particle, isolated from a patient. Image captured and color-enhanced at the NIAID Integrated Research Facility (IRF) in Fort Detrick, Maryland. Credit: NIAID
Comparing CoV and influenza viruses
The current study compares the genome of SARS-CoV-2 to that of the influenza virus to explore its transmission and whether it can pose a future threat. The researcher used a technique called principal component analysis.
The influenza virus and SARS-CoV-2 virus are similar in many ways. Both are RNA viruses. Both replicate via RNA-dependent RNA polymerases, which are sources of genetic variation leading to the formation of different classes and subclasses of the virus.
However, they are also different in the specificity with which they infect hosts, and how fast they mutate.
No intermediate host for CoV?
For instance, many bat coronaviruses (CoV) can infect humans directly, without the need for an intermediate host. This character may have helped jump species barriers from bat to human. This phenomenon may also be a reflection of lower selection pressure due to the lower rate of infection.
Every coronavirus that caused a human outbreak has a similar counterpart in bats or camels, with no unique residues in the coronaviruses that cause COVID-19. Even with SARS, only a small minority were different, with many showing variations even among the bat CoV.
Bat viruses undergo spontaneous variation as a result of RNA editing and replication errors, so multiple variants are present even in bats. This makes the border between bat and human viruses very thin, unlike what would be expected from in vitro studies because of the high tendency to variation, which allows adaptation to occur once an infection is established.
Intermediates are not, therefore, strictly necessary as cumulative adapting mutations need not occur to infect humans. Even if they are needed, their primary role could be to amplify the dose of the virus needed and the contact frequency.
To avoid the development of intermediate hosts, the authors advise that live animals be kept away from infected animals. People who frequently come in contact with wild animals should not be allowed to live in a large city. Early cases should be identified to prevent a human outbreak.
Rapid mutations
The coronaviruses as a whole do not vary as much or as fast as the influenza viruses do. The coronaviruses that occur among humans are different. They quite often undergo annual mutations (like the influenza H virus).
If this occurs every year, its highly infectious nature coupled with genomic changes can cause repeated outbreaks to spread every few years. Some kinds of CoV cause infection of the upper respiratory tract in humans, like the common cold, but also severe pneumonia.
The coronavirus quiver is not yet empty. There are many other CoV that can cause human infection. Like the influenza virus, the coronaviruses are RNA viruses, but with a single strand of RNA in contrast to the eight segmented influenza genome.
They are approximately 15 times less infectious, on average than the influenza viruses. However, some like the MERS or SARS-CoV and the present virus are much more infective than the older ones and thus cause outbreaks.
Middle East respiratory syndrome coronavirus - Image Credit: European Centre for Disease Prevention and Control (ECDC)
Classes of CoV – genetic divergence
SARS-CoV and SARS-CoV-2 are Sarbecovirus subclasses. However, the researchers found that Alphacoronavirus and Betacoronavirus classes were wide also. Gammacoronavirus and Deltacoronavirus are chiefly bird CoV, while mammalian CoV seems to come from the bat CoV (Norbecovirus).
Human coronaviruses (HCoV) are in the Embecovirus, Alphacoronavirus, and Dubinacovirus classes, while the MERS and SARS CoV belong to more recent classes, the Merbecovirus and the Sarbecovirus, respectively.
All these contain many insertions and deletions (indels) of smaller open reading frames. The different classes are close together in the genetic variation graph. Influenza viruses, however, are widely separated depending on the host. There is a magnitude of difference between the divergence in coronaviruses compared to that in the influenza viruses.
Breaching species barriers
The researchers say, “Coronaviruses can apparently breach cell type, tissue, and host species barriers with relative ease.”
The reason could be that the CoV spike protein is more adaptable to host differences than the influenza virus hemagglutinin protein. This could reduce the selection pressure and thus the divergence.
CoV show shifts, but not influenza viruses, because of the latter’s ability to replace RNA molecules between viruses. The CoV cannot transcribe the genome into double-stranded DNA, which means it uses other mechanisms. These could include nuclear RNA splicing or RNA interference, which can both cause frequent indels.
Why does genetic variation occur?
Virus genetic variation is due to several selection pressures, to maintain their function by avoiding nonfunctional mutations; to preserve infectivity in the face of rapidly spreading host immunity, by constantly mutating like Influenza A; and to infect new hosts by alterations in the cell receptor attachment apparatus as well as genetic adaptations to the host; and finally, to induce a large number of asymptomatic infections and thus avoid containment.
Will CoVs cause future pandemics?
Unlike the earlier coronavirus outbreaks, the SARS-CoV-2 has the conditions necessary to cause a pandemic, like the influenza viruses. It is quite infectious. It does not induce herd immunity rapidly. It is not subject to large selection pressures.
If so, it is quite feasible that the virus will persist among humans and continue to cause spreading outbreaks every few years. Already, SARS-CoV-2 has given rise to 30 strains and multiple classes within months of the onset of the pandemic.
The changes could cause better adaptation to humans, reduce herd immunity, or reduce the fatality rate. However, the CoV have shorter lifespans than influenza viruses. They also have smaller open reading frames (ORFs), which means reduced variability.
This could, therefore, be an excellent target to induce herd immunity and for vaccine development, as they are conserved in most CoV. The spike protein of CoV is too variable and could cause enhanced undesirable effects in the presence of antibodies, on the other hand.
The authors sum up: “If SARS-CoV-2 changes its genome like the Influenza H type, it will repeatedly spread every few years. In addition, the coronavirus family has many other candidates for subsequent pandemics.”
Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.