The transmembrane protease serine-type 2 (TMPRSS2) protein plays a key role in COVID-19 infection since it primes the viral spike protein to allow viral entry into the target cell. A new study by researchers at Imperial College London and published on the preprint server bioRxiv* in May 2020 describes the protein, which could be an attractive drug target to help manage COVID-19.
The function of the TMPRSS2 includes the ability to cleave and activate the spike protein of several coronaviruses, including the SARS-CoV, which caused the earlier 2002–2004 SARS outbreak. This cleavage is required for viral-cell membrane fusion and viral infection. This protein is among the main proteases on the cell surface that take part in this priming process, in addition to furin and lysosomal cathepsin.
The reason for the broad spectrum of symptoms in COVID-19 is unknown. There is little known variation in the ACE2 gene. Could it be due to genetic TMPRSS2 variants?
Earlier studies have shown that these coronaviruses cannot replicate in the absence of this enzyme, causing a reduced immune response as well. Inhibitors of this enzyme also prevent bronchial cell infection by SARS-CoV in vitro, an effect confirmed with protease inhibitors in animal studies.
Based on these results, the current study aims to explore the putative protective effect of natural genetic variants that alter the structure and function of the TMPRSS2 protein. The GnomAD database of genetic variations in the population was analyzed using computational bioinformatics to identify those variants that affect protein structure and function, as well as to find how commonly they are distributed in the population.
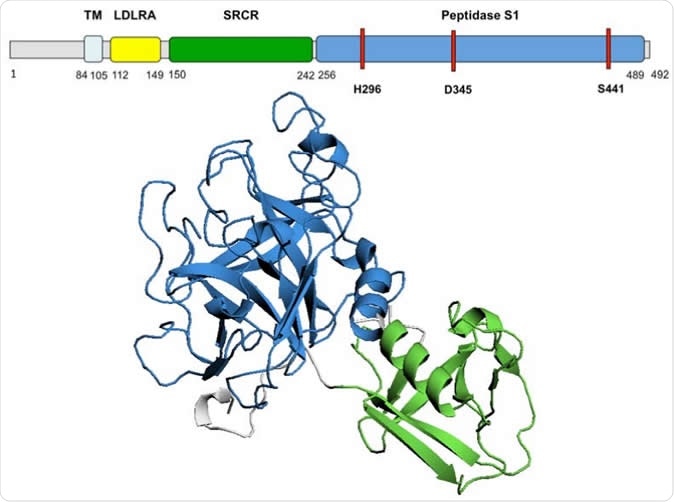
TMPRSS2 predicted 3D structure Diagram of TMPRSS2 amino acid sequence and domains. The 3D model of TMPRSS2 domains SRCR and Peptidase S1 is presented. The active site, residues H296, D345 and S441, is highlighted in red on the amino acid sequence. TM, transmembrane domain; LDLRA, LDL-receptor class A; SRCR, scavenger receptor cysteine-rich domain 2; Peptidase S1, Serine peptidase.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
The researchers also used human protein atlas data to find out whether ACE2 and TMPRSS2 were co-expressed in extrapulmonary tissues, especially the gut, because of the common occurrence of diarrhea and other gut symptoms in COVID-19.
As a first step, they built a 3D model using a Phyre modeling algorithm and evaluated its quality with three different methods. They also assessed the impact of each of the variant forms on the structure of the protein using the Missense3D algorithm, as well as other methods.
The researchers found that the TMPRSS2 protein is built of a cytoplasmic region, a transmembrane region, and an extracellular region. This last region has three domains, of which the peptidase S1 contains the active protease site, which can be glycosylated at 2 positions, along with a cleavage site that allows the extracellular region to be shed.
The 3D structure of the extracellular region was found to be of good quality. Of the 378 TMPRSS2 variants analyzed in the current study, one variant, p.V160M, was found to have a minor allele frequency (MAF) of 0.248 in the population, slightly higher among males compared to females.
This corresponds to a distribution of about 25% of the general population. About 6.7% of the population was homozygous for this variant, which has been predicted to be a damaging substitution by all the methods used.
The SCRS domain is highly conserved, but its function is not entirely clear. It may be necessary in ligand or protein interaction. It is seen in several host defense proteins, which could indicate a role beyond its protein-cleaving activity that primes the virus for membrane fusion and viral entry.
Within this SCRS domain, valine 160 is a small but highly conserved amino acid which could have an important role in the structure or function of the protein. Though the steric clash was suspected, it was not observed to be likely from the change in free energy.
There are 31 variants that could stop the translation of the protein prematurely. Among 304 variants which were mapped on to the 3D structure generated by the researchers. Of these, 62 are thought to be structurally damaging according to the MissenseD prediction, and 12 are highly destabilizing to the protein, possibly leading to protein structure misfolding.
Two of the variants disrupt protein function as well, namely, p.R255S and p.S441G. The first of these abolishes the TMPRSS2 cleavage site while the other abolishes the active site of the protein. The fact remains that their rarity in the population reduces their utility as markers of severe COVID-19 infection.
Another 167 or 152 variants were predicted to be damaging, depending on whether the SIFT or Polyphen programs were used. However, 137 of these were common to both. Structural damage was predicted for 53 of them, which could indicate that they cause severe damage to the protein.
Overall, the 53 structural damage variants and the 31 prematurely truncated protein variants are predicted to probably cause loss of function but are infrequently distributed in the population. Both the mean and the cumulative MAF are low, meaning that their use as markers of severity of SARS-CoV-2 infection is unlikely.
Are TMPRSS2 and ACE2 Co-Expressed in The Intestine?
The data on TMPRSS2 and ACE2 expression taken from the Human Protein Atlas showed co-expression of both proteins in the tissues of the gastrointestinal tract, the kidney, and the gallbladder. This could indicate the susceptibility of the gut to this virus, explaining the gut symptoms often seen in COVID-19 infection.
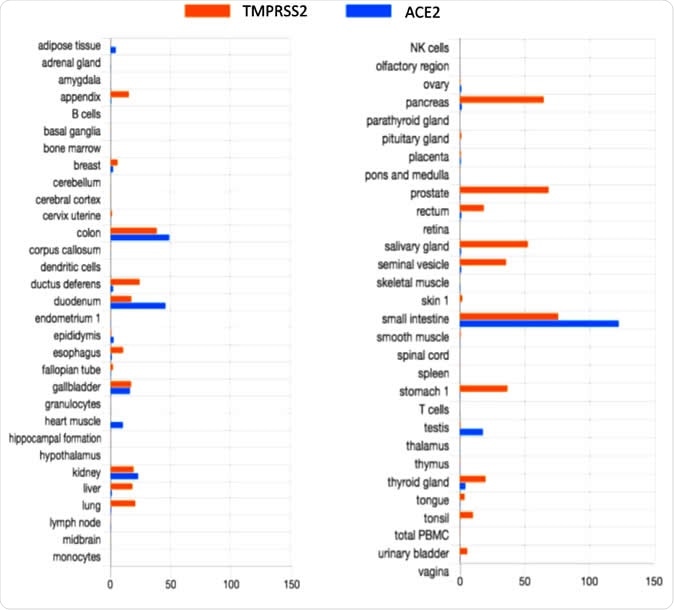
TMPRSS2 and ACE2 tissue expression
However, the HPA fails to show the expression of ACE2 in the lung cells, or in the endothelial cells, or arterial smooth muscle cells. This is despite the fact that recent work has shown the co-expression of both genes in lung and bronchial tissue. This indicates the need for specific in vitro experiments to examine this feature in the gut and other tissues as well, completing the HPA data.
According to the researchers, the study shows that “TMPRSS2 variants should be investigated further to understand the impact of a person’s genetic background on their clinical presentation and prognosis when contracting SARSCoV-2.” Further studies on the co-expression of these gene variants in the cells of the gastrointestinal tract are also required.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.