The COVID-19 pandemic has left its mark on India after it popped up in China at the end of 2019 and spread to over 188 countries and territories within just a few months. While non-pharmacological interventions have been the mainstay of governments and health organizations in dealing with the virus, in the absence of effective drugs or vaccines, the virus has not yet been tamed.
The Early History of the Pandemic in India
The first three cases in India cropped up in the progressive southern state of Kerala, all in Wuhan returnees. With the immediate quarantine of these cases, no local transmission occurred. However, India also introduced steps to prevent further introduction of cases, blocking flights from affected countries. In March, several new cases cropped up as a result of importation from other countries, with associated local transmission.
This was followed by a national lockdown from March 25, 2020, in an attempt to check the spread of the virus. However, with partial relaxation of these measures in the early part of May, the number of cases began to rise by leaps and bounds as people crisscrossed India in a frantic attempt to get home across newly reopened state borders.
Tracing Genomic Evolution
At present, there are over 230,000 confirmed cases in India, as of June 6, 2020. To understand the origin of the strains causing this epidemic, the researchers performed whole-genome sequencing (WGS) of 104 strains of SARS-CoV-2 from all over India. They retrieved genomic data from the Integrated Disease Surveillance Program (IDSP) of the National Centre of Disease Control (NCDC), Delhi.
Using both genetic and epidemiological data, the study helps uncover the breadth, evolution, incidence, distribution as well as control of COVID-19 in India. This is expected to help with the contact tracing, as well as the development of diagnostics and therapeutics for the disease.
The study was carried out by the NCDC along with the CSIR-Institute of Genomics and Integrative Biology (CSIR-IGIB). The researchers included 127 confirmed cases from different locations, identified by targeted testing of symptomatic individuals who had a history of travel to high-risk locations or contact with COVID-19 patients.
The nasopharyngeal and oropharyngeal swabs were used to obtain viral RNA for WGS. The mean age was 41 years, with the male: female ratio being 35:28 below 39 years, and 58:6 above this age.
Most of the samples came from New Delhi, with a few coming from clusters in various other states. While the majority were Indian, 14 were Indonesian, and 2 each from Thailand and the Kyrgyz Republic.
The Study Results
There were 104 samples that passed quality testing and were used to map the complete genome. The phylogenetic tree constructed from these strains showed that all the strains were grouped into 2 major clades, with a few other miscellaneous clades and a sub-clade. Overall, there were 163 variants, of which less than 5% were common.
One cluster of 26 belonged to Cluster 1, in the G-clade as classified in the Global Initiative on Sharing All Influenza Data (GISAID). Another cluster of 65 strains belonged to Cluster 2, which is an unclassified cluster according to GISAID. Most of the important variants in this cluster are also seen in sequences from Singapore, Brunei, and the sequences from India came from Indonesian, Thai and Kyrgyz natives to a significant degree, besides those from Tamil Nadu and Delhi.
The researchers comment, “This probably suggests the introduction of this particularly from East Asian countries into India.”
Cluster 3 has 7 strains that segregate with the other strains from India. There were 2 belonging to A1a, and 3 from B, with a Maharashtra strain that showed no variants, and is probably identical to the original Wuhan strain.
Mutations and Protein Effects
The investigators also looked at the mutant proteins expressed as a result of amino acid substitutions across the 104 genomes. There were 53 point mutations, of which 29 resulted in missense mutations.
When studied in relation to the proteins affected, the scientists found that most variations were in non-structural protein (nsp) 6, in 68 genomes, with nsp 12 in 65, nsp 3 in 62, and P13L, a nucleocapsid protein, in 53. One of the commonly found worldwide mutations, D614G, in the spike protein, was found in only 26 genomes.
The researchers then followed the frequency of mutation with respect to the type of change in the amino acid. They found that in about 45%, the amino acid was not changed, which indicates that perhaps the mutation caused a slight change in the shape or function of the protein. The same thing was observed with very frequently found mutations, such as P13L.
In some other mutations, the type of amino acid was quite different, with a hydrophobic-to-polar or charged type of alteration. One such example is that of the addition of a charged residue to the frequently mutated position T1198K in nsp3, or the loss of a charged group with the important spike protein mutation D614G. Such a changeover to a residue with a positive charge may possibly result in more significant effects on the structure and function of the protein involved.
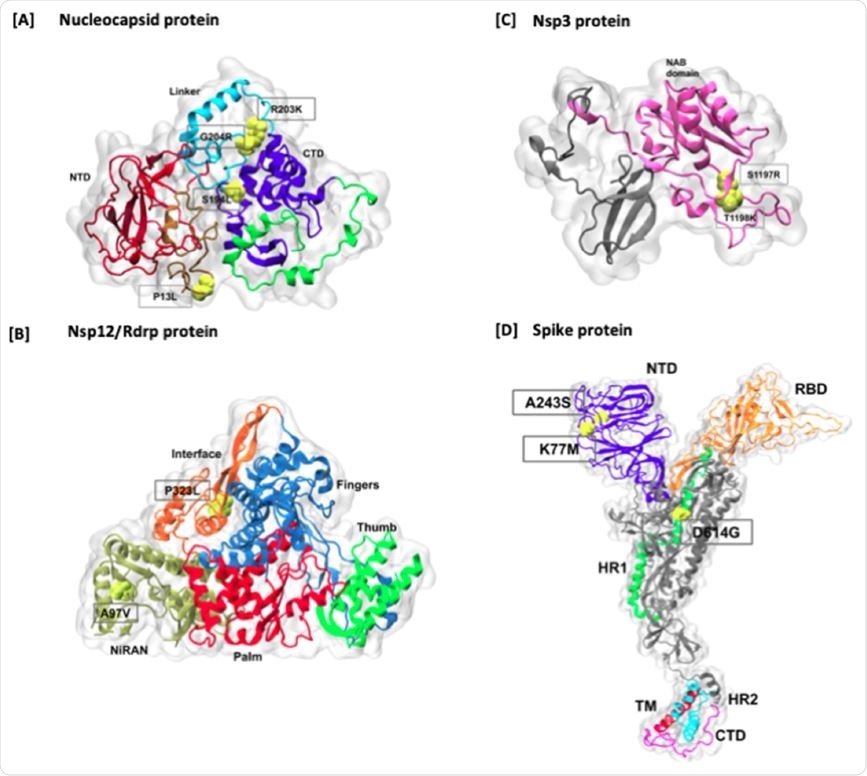
Mapping of higher frequency amino acid mutations on Nucelocapsid and Spike proteins. The mutations are marked in red color on the surface representation of each protein. In Spike protein, all the domains are highlighted in different colors, including NTD, RBD, HR1, Fusion peptide region, HR2, TM, and CT domains. In addition, cleavage sites are also marked onto the structure.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Mutations and Local Environment
The sites undergoing mutation in SARS-CoV-2 were compared with six other Coronavirus sequences and found to be mostly in variable locations (19/29 mutations). The mutations occurring at a higher frequency are in positions that change faster, except for two at conserved locations, namely, A97L and L37F.
The effect of these locations on the local environment is an important aspect of such a study. To understand this, the scientists traced the link between the most common mutations and the viral protein structures. They found that all the mutations occur outside the two structural C- and N-terminal domains in the nucleocapsid, and in the longer linker regions.
Mutations in nsp12, which is highly conserved, are on the interface region, which has an essential zinc-binding site. On the other hand, the P323L mutation is at the protein interaction junctions, where inhibitors bind to a hydrophobic cleft and its replacement of proline with leucine results in the loss of the kink at this site. Similar mutational effects on the protein products are seen in nsp3 and the spike protein.
Three Waves of Invasion
The researchers describe the three waves of viral entry into India, first from European and American travelers, the second from the Middle East, and the third from South-East Asia. They found that the A4 cluster, though not classified so far, is the most prevalent among Indian genotypes.
They also identified some novel mutations, but a more thorough evaluation is necessary to validate these findings.
The study also reveals the failure of the lockdown because of the shift of cases from mostly urban locations to rural areas mediated by the vast exodus of migrant laborers from multiple states in India to their home states. The expected community transmission as a result of this movement will require energetic preventive measures.
The large-scale lockdown may have led to the preferential evolution of certain strains that easily adapted to Indian conditions, and this may have resulted in the emergence of a distinct lineage. This still awaits addition to international viral genomic databases like Nexstrain.
Future Directions and Implications
A more rigorous study will determine if these strains are to be included in designing diagnostic tests. If so, it may enable more cost-effective panels to track the spread of different lineage-specific strains crossing geographical boundaries with greater speed and effectiveness.
Vaccine development must also consider the prevalent variants in India. Moreover, relating the prevalence with the clinical history will allow identification of those strains that are most pathogenic and virulent in terms of causing severe disease.
The researchers conclude, “It is imperative that robust genomic data based on large sample size including rural populations with even distribution can bring out the real scenario once correlated with epidemiological data eventually helping in the drafting of further management policies.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Kumar, P., et al. (2020). Integrated Genomic View Of SARS-Cov-2 In India. bioRxiv preprint. doi: https://doi.org/10.1101/2020.06.04.128751
- Peer reviewed and published scientific report.
Kumar, Pramod, Rajesh Pandey, Pooja Sharma, Mahesh S. Dhar, Vivekanand A., Bharathram Uppili, Himanshu Vashisht, et al. 2020. “Integrated Genomic View of SARS-CoV-2 in India.” Wellcome Open Research 5 (August): 184. https://doi.org/10.12688/wellcomeopenres.16119.1. https://wellcomeopenresearch.org/articles/5-184/v1.