A preliminary study by researchers from the Garvan Institute of Medical Research and the University of New South Wales and published on the preprint server bioRxiv* in June 2020 reports the use of chimeric design to produced reference standards for diagnostic tests. This could allow a highly flexible approach to the development of different genetic features in the same test, even for organisms that are not directly related.
With the continuing pandemic of COVID-19, diagnostic and therapeutic measures are still being intensively developed. Concerning diagnostic tests based on the detection and quantification of viral RNA or DNA, reference standards are a must. DNA synthesis is a new technology used to create synthetic molecules that act as the target for genetic detection methods.
These molecules can be used to rapidly-produce a lot of reference standards, and can even incorporate sequences from other organisms to meet the needs of a particular assay. The current study uses this method to create artificial reference sequences that contained the primers for all the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) assays currently published by the World Health Organization.
These synthetic references can be used as positive controls, with their reaction showing the sensitivity of the molecular test being evaluated. On the other hand, negative controls that lack the target sequences must also be used to identify the specificity of the test.
The Problems with Natural Constructs
Any reference must be first validated and shown to be suitable for the purpose of the test. With RNA sequences, the control constructs are bound to break down over time, and contamination is also possible, both of which can lead to confusing test results. The possibility of failure could be on the part of either the test, the control, or the method used. Such failures cause diagnostic delays, misdiagnosis, and the non-acceptance of valid test results.
Synthetic Chimeric Constructs
For this reason, RNA and DNA standards are now being synthesized in vitro, to make the design process more flexible and thus offer tailored controls that fit the diagnostic test and the targets. The current paper reports a new strategy for designing reference standards that use chimeric synthetic standards matched to each other to meet the standards of a testing methodology called A/B testing.
The target sequences of a molecular test are first obtained and divided into two groups A and B. These are then joined end-to-end to get a single chimeric sequence. The advantage of equally splitting the target sequences between the two groups is that each target has its positive control in one group and negative control in the other group, with positives and negatives free to be in either group.
This type of equal partitioning allows the controls to validate each other, making the test results more dependable and allowing the failure of a control to be distinguished from a failed or successful test. Thus, the target regions of a single organism can be tested together, even if they are quite different. The same applies to target regions from different organisms.
The Current Approach: Chimeric Pairs for Validation
Using this approach, the researchers used a pair of chimeric A/B standards to carry out the RT-PCR diagnostic testing for SARS-CoV-2, according to the real-time tests recommended by the World Health Organization (WHO). Both the standards had regions of the viral genome that are recognized by published primer pairs, and are compatible with already licensed and approved diagnostic tests.
The current study looked at the performance of these controls in comparison to other reference sequences and clinical specimens, showing how these synthetic constructs can be used to validate both the RT-PCR and also showing how they can be used in other tests as well.
The initial step was to obtain the viral genome sequences for the initial isolate and all WHO-published primer sequences for six countries, 3 in Asia, 2 in Europe, and the USA. Each test now available uses two or three pairs of primers, directed against various parts of the genomic sequence. These were aligned to the whole genome so as to identify the locations of the amplified sequences. These amplicons were retrieved with 30 nucleotides belonging to each end.
This design allows the use of the chimeric standards in each country separately as test controls, as well as a common standard to enable the direct comparison of test results and efficiency in different sets of primers and probes and countries.
These retrieved sequences were next split across two control groups, namely, chimeric A/B standards. The country-based target regions were assigned to two different groups each, after which the regions were assembled in tandem. Chimeric standard B also contained a sequence from the human RNAse P (RP) gene, used as a positive human control.
So also, the standards contained a unique control sequence to help the detection of each of them, if necessary. Each had an initial promoter region so it could be transcribed in vitro, with a terminal poly-A tail and a restriction site, to allow vector linearization.
The two different A/B standard sequences were then synthesized. The product was cloned to plasmids, which were then linearized and transcribed in vitro. The final product was the synthetic RNA standards, after purification and validation.
They then used other reference controls to validate the chimeric standards and tested them against genomes from 12 COVID-19 patients, which had already been independently sequenced. As expected, the RT-PCR diagnosed both chimeric standards and patient samples, with target genes N1, N2, and N3 being amplified within patient RNA and patient cDNA.
The researchers found that both standards could be detected until 10-4 dilution or a limit of detection for a copy number of 0.39 and 0.42 copies/μl for standards A and B, respectively.
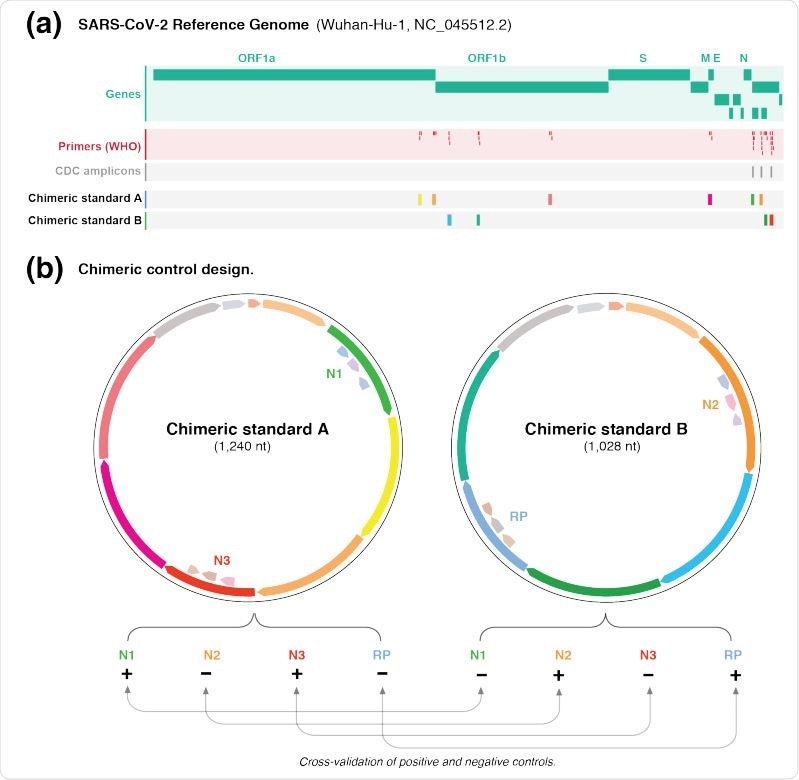
Design of chimeric controls for SARS-CoV-2. (a) Genome browser view of the SARS-CoV-2 genome (green) showing WHO-published real-time RT-PCR primer binding sites (red). The expected amplicons for the CDC test are shown in darker grey. The other targeted regions were exclusively partitioned between chimeric A/B standards. (b) The different targeted regions for standards A and B were shuffled and joined together to form chimeric sequences. The paired design of chimeric A/B standards, where a target in A is absent in B (and vice versa), enables the synthetic RNA transcripts to simultaneously act as positive and negative controls for the real-time RT-PCR primer/probe sets. this enables internal cross validation of positive and negative controls between standards A and B. The vector backbone was omitted from the representation of the chimeric A/B standards.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Future Applications
The applications for this technology include not only novel diagnostic technology but also testing for multiple genetic features like mutations and viral sequences, cancer mutations, and gene signatures. Secondly, when the current pandemic begins to decline, such controls will be required for accurate and rapid diagnosis of infection.
The study sums up: “The chimeric A/B approach provides a new model for the development of cost-effective reference standards that allow for controlling of multiple experimental variables based on simple, but comprehensive designs.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources