The urgency of finding a solution for the ongoing COVID-19 crisis has led to the sequencing of hundreds of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genomes, to help understand how the virus invades human cells, how it replicates and causes disease and factors that help it evade immune defenses.
Novel Coronavirus SARS-CoV-2 Colorized scanning electron micrograph of a VERO E6 cell (blue-green) exhibiting elongated cell projections and signs of apoptosis, after infection with SARS-COV-2 virus particles (yellow), which were isolated from a patient sample. Image captured at the NIAID Integrated Research Facility (IRF) in Fort Detrick, Maryland. Credit: NIAID

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Antiviral Defenses and Coronaviruses
Coronaviruses (CoVs) infect mammals, and their genomes are shaped by their environment, which varies with the host tissue. This is because different species have different kinds of cells, with significant differences in the antiviral and RNA modification level of activity.
One important component of the antiviral response is the production of antiviral protein (AVP), among which is the zinc-finger antiviral protein (ZAP), encoded by gene ZC3HAV1 in mammals, and the apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3 (APOBEC3).
ZAP is part of the interferon-mediated immune response, which is directed against CpGs in the viral genome, leading to the inhibition of viral replication and the production of the signal to break down the viral genome. The presence of ZAP activity in the cytoplasm should cause a viral genome to avoid CgPs if the host tissue is abundant in ZAP, as seen with HIV-1, and many CoVs. HIV-1 virus is lymphotropic (lymph nodes being ZAP-enriched) but also CgP deficient. With increasing CgP content, the HIV-1 becomes less fit.
ZAP causes increasing and cumulative degradation of viral RNA, as the number of CgPs increases all through the RNA sequences. Concerning SARS-CoV-1, this would mean that ZAP will target only ORF1 and Spike mRNA, which are of sufficient length. Even with its relatively low CpG content, among betaCoVs, these mRNA sequences have few CpGs.
The APOBEC3 is also important in the antiviral response. They cause hypermutations in the viral cDNA, so that the transcription is defective, resulting in the inhibition of reverse transcription. It can bind to RNA and thus become incorporated into virions. It also edits C to U in RNA.
The HIV-1 prevents these actions through its Vif protein that degrades this enzyme. Nonetheless, this may present the potential for the development of paralogous drugs that can directly edit ssRNA viruses that lack the Vif analog – like SARS-CoV-2.
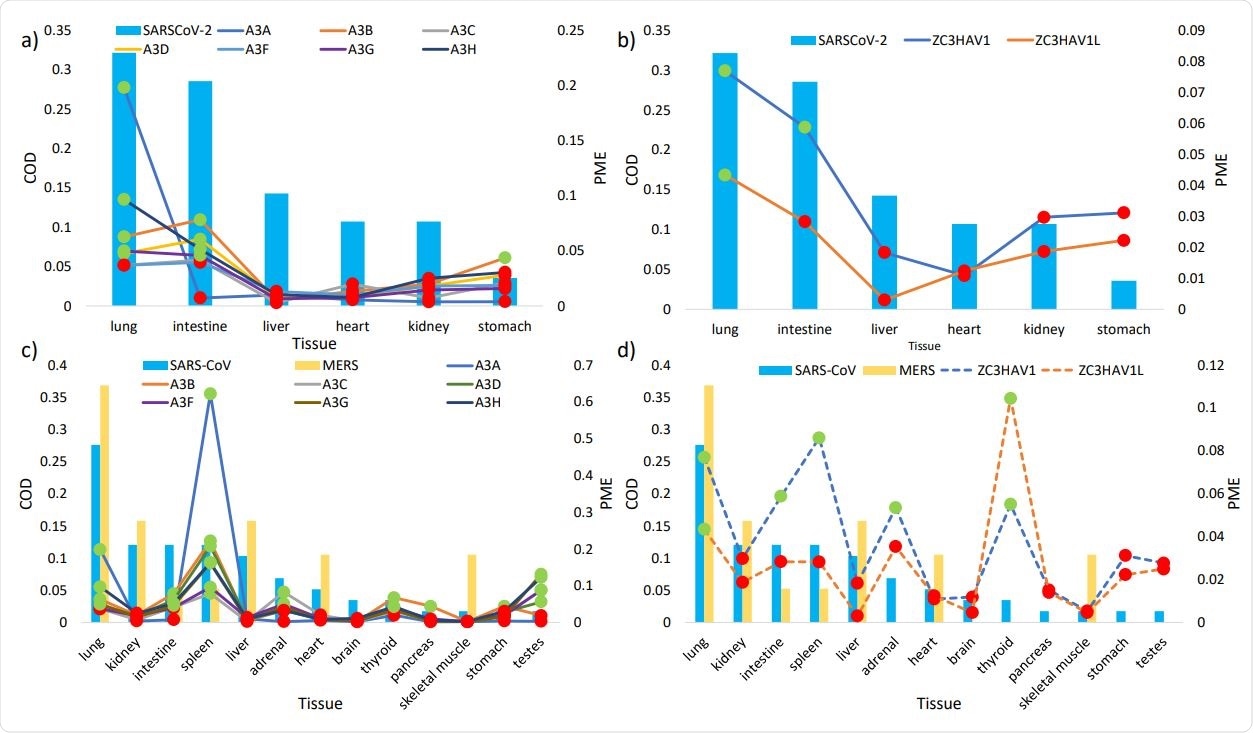
The histograms show the regular tissue habitats (as measured in commonness of detection COD, on primary Y axis) of SARS307 CoV-2 (a, b) and of SARS-CoV and MERS (c, d). The lines represent the relative mRNA expression (in proportions of mRNA expression PME, on secondary Y axis) of a) APOBEC3 isoforms (solid lines) and b) ZAP isoforms (dash lines) in tissues susceptible to SARS-CoV-2 infection, and the PME of c) APOBEC3 isoforms (solid lines) and d) ZAP isoforms (dashed lines) in tissues susceptible to SARS-CoV and MERS infections. Highlighted in green and red are PME values that are greater and lower than the averaged PME values, respectively. PME values were calculated based on averaged mRNA FPKMs retrieved from the GTEx Portal (LONSDALE et al. 2013).
Aims of the Study
The current study aims at testing the hypothesis that both these factors act together as the primary source of selective pressure that drives the CoV to adapt as it infects different host tissues. The researchers analyzed the antiviral proteins that are effective against CoV and the mechanism of suppression of the immune response.
They predicted that the specific host tissue environment could be identified depending on the presence of consistent CgP avoidance or U enrichment. These changes would mark the presence of abundant antiviral proteins (AVP) such as ZAP or APOBEC3 in the host tissue typically infected. On the other hand, their absence predicts the typical infection of host tissue lacking AVP.
The researchers included a large number of publicly available genome sequences for seven CoVs, as well as specific patterns of these AVP in different tissues among five host species – humans, dogs, pigs, mice, and cattle.
Host Tissue AVP Direct Viral Genome Changes
The study shows that these viruses (except for one, the murine MHV) typically infect tissues with high AVP. Concordantly, they all have high deficiencies of CpG throughout their sequences, with SARS-CoV-2 having the lowest CpG content, which is spread across 12 regions of the genome in a non-uniform manner.
The lowest CpG content is in the region that encodes the S protein. This pattern indicates the emergence of the virus in a host tissue with high ZAP levels and that this has helped it to avoid the antiviral defenses triggered by ZAP during its entry into the cell. Thus AVP-rich host tissues produce a shift towards CpG deficiency and higher U content in the viral genomes.
Evidence of C to U switch at the level of RNA, which is brought about by APOBEC3, is found in many CoVs. However, the proportion of U is comparable to that of murine MHV though the latter does not infect AVP-rich tissues, So, does APOBEC3 play a role in shaping the genome of those CoVs that infect human hosts?
To answer this, the researchers looked at the patterns of single nucleotide polymorphisms (SNPs) in the local regions of various genomic sequences for the SARS-CoV-2 from the four months (December 31, 2019, to May 6, 2020).
The researchers found that the cumulative elevation in U and decline in C content is not remarkable in the SARS-CoV-2, SARS-CoV, and MERS, because of the insufficient time to accumulate a large number of mutations in these recently emerged strains.
On the other hand, C to U substitution is far more prevalent than other SNPs, and this is seen mostly in the 5’ UTR and the ORF1ab regions, indicating that these are under selective pressure by the AVP. When 99 complete genomes collected on different days over this period were analyzed, the number of C to U substitutions was seen to be higher than expected, and increasing over time, in these two regions alone.
In other words, APOBEC3 also plays a role in directing the evolution of this virus.
Other types of change, such as the A to G mutation, are seen to occur in the S region, perhaps due to the activity of the mammalian ADAR1 enzyme. However, the CpG content declines slowly over time, as shown by the lack of difference between the samples collected over four months.
What Implications Does This Carry for SARS-CoV-2?
An important implication of this analysis is that the SARS-CoV-2 became CpG deficient in an intermediate reservoir before it jumped to humans since this change does not take place rapidly. The sequencing of the bat CoV RaTG13 back in 2013 would have warned of the high probability of the virus shifting to a host with high ZAP expression, since it had successfully changed to avoid this antiviral defense.
Another conclusion is that the evolutionary pressure targets CpGs in specific regions, focusing on the region that is vital for host cell recognition and entry, namely, the spike protein.
The researchers suggest that the possibility of increasing CpG content, which is known to reduce viral replication and virulence sharply, is one to be explored while developing weakened SARS-CoV-2 strains for vaccine development. The counter side is that this may be subverted by host innate deaminases, which change CpGs into UpGs, further increasing the level of CpG deficiency. Thus, more research into how AVP act to direct the evolution of the virus is required.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources