The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that is causing the COVID-19 pandemic across the world has undergone a genetic mutation in the density of functional spikes on the virus, says a new study. This has led the virus to become highly infectious and easily transmissible between humans.
What was the study about?
The SARS-CoV-2 has successfully infected over 8 million individuals across the world and led to over 435,000 deaths, as of 16th June 2020. The virus body is covered with spikes that give it its name “corona,” which means “crown” in Latin. These spikes are essential for the virus to get attached to the ACE2 (angiotensin-converting enzyme) receptors on the surface of the host cells. From these entry points, the virus can successfully enter the host cells and infect the individual. The virus uses the host cell machinery to replicate and spread to other cells causing the infection COVID-19.
The SARS-CoV-2 isolates have been found to have a minuscule mutation called the D614G mutation. This mutation is seen on the viral spike protein, also called S-protein. This research attempted to compare the functional properties of the S-protein with other proteins such as “aspartic acid (SD614) and glycine (SG614)”. These other two proteins are at residue 614 on the viral genome, the experts said.
What was found?
In their study, the researchers noted that if they caused other retroviruses to have the SG614 mutation, they were more capable of infecting cells that contained the ACE2 receptor compared to those with SD614 mutation. They noted that with this mutation, the S-protein was incorporated in the artificial viron (pseudovirion). There was less shedding of S1, they wrote.
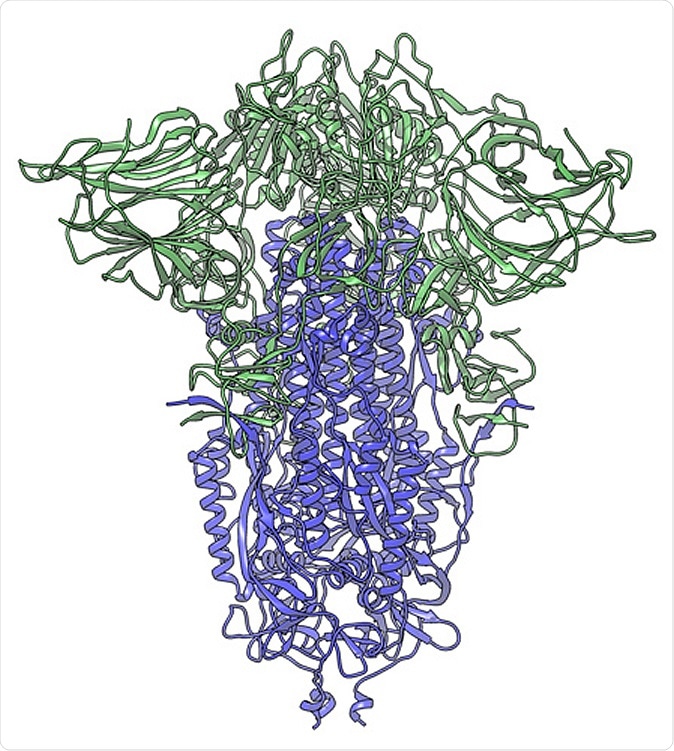
In this cryogenic electron microscope image of a SARS-CoV-2 spike protein side view, the S1 section of the spike is shown in green and the S2 portion is shown in purple. This unique two-piece system has shown itself to be relatively unstable. A new mutation has appeared in the viral variant most common in New York and Italy that makes this spike both more stable and better able to infect cells. (Credit: Andrew Ward lab, Scripps Research)

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Scripps Research virologist Hyeryun Choe, Ph.D., senior author of the study, said, “Viruses with this mutation were much more infectious than those without the mutation in the cell culture system we used.” She explained that this mutation allowed the virus to have more functional spikes on its surface and thus allows it to stick with the host cells more than before. She added, “The number—or density—of functional spikes on the virus is 4 or 5 times greater due to this mutation.”
Co-author Michael Farzan, Ph.D., co-chairman of the Scripps Research Department of Immunology and Microbiology, added in explanation that the mutation D614G could cause improved flexibility to the basic backbone of the structure of the S protein. This can allow it to latch on better with the ACE 2 receptors and thus cause the infection. This flexibility of the spike can allow the replicated viruses from the initial host cell to move unharmed to the next target cells and thus infect multiple cells. Those without the mutation do not have a stable S protein, which allows them to break easily while replicating and moving on to secondary target cells.
Choe added, “Our data are very clear; the virus becomes much more stable with the mutation.” Farzan said, “There have been at least a dozen scientific papers talking about the predominance of this mutation. Are we just seeing a ‘founder effect?’ Our data nails it. It is not the founder effect.” What they meant was that their biochemical experiments provide the necessary proof that a small number of mutated viruses or variants have spread to a wider population called the “founder effect,” which was not seen.
The team had also produced virus-like particles using SARS-CoV-2 M, N, E, and S proteins. The results with the mutation remained the same, they wrote. SG614 was found to not bind effectively with ACE 2 compared to SD614. In the pseudoviruses created, the S-proteins were found to be neutralized when exposed to antibodies found in plasma from patients who had already recovered from COVID-19 (convalescent plasma).
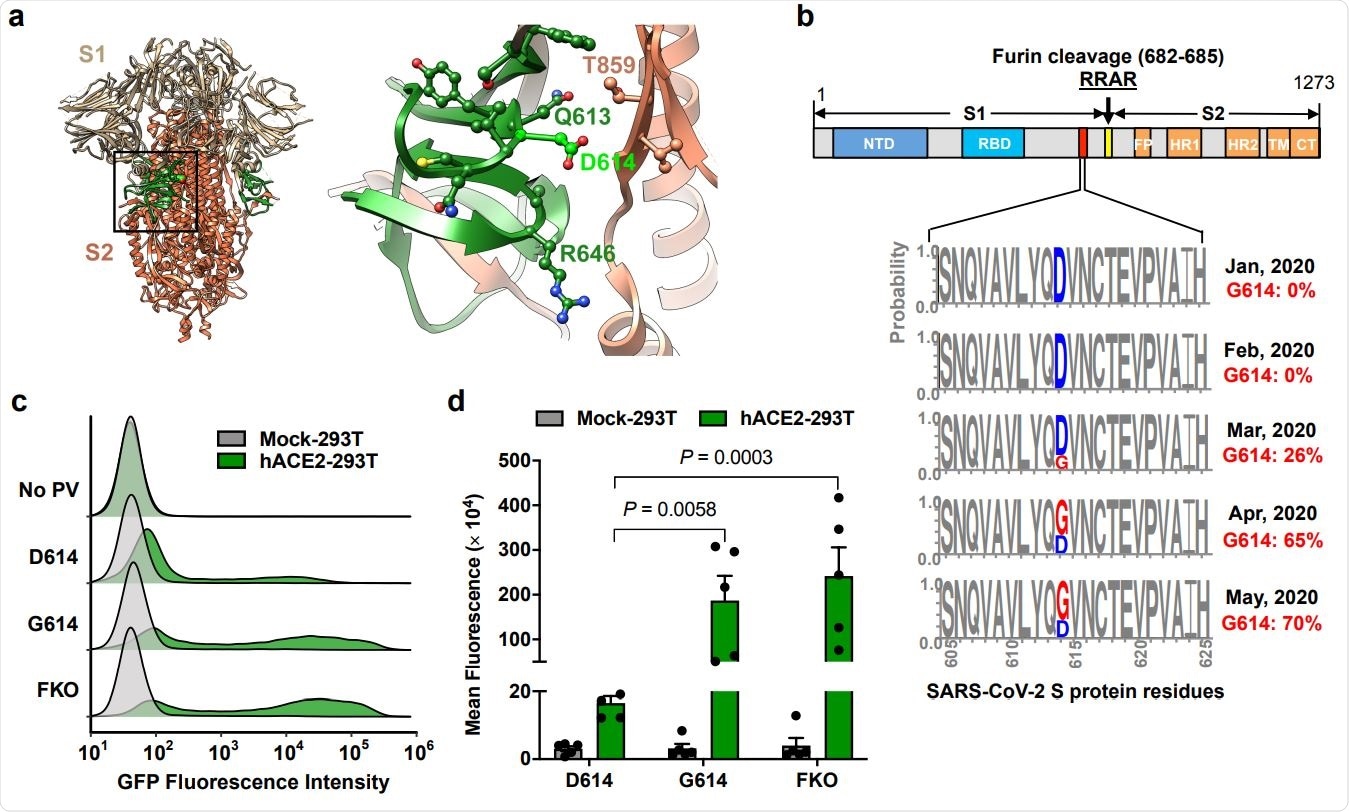
The D614G mutation is associated with enhanced infectivity. Cryo-EM structure of S1 (grey) and S2 (orange) heterodimer (PBD 6VXX). The residues 581-676, a C-terminal region of the S1 domain involved in S2 interaction, is shown in green. Aspartic acid 614 is shown in light green. The area indicated with a black square is presented magnified at the right. Residues within 5.5 Å of D614 are shown in a ball-and-stick representation. b, A representation of the SARS-CoV-2 Sprotein (upper panel) and D/G variation at the residue 614 presented in logo plots at different time points between January 1st and May 30th, 2020 (lower panel). The total number of sequences analyzed: 17 on January, 33 in February, 293 in March 1511 in April, and 2544 in May. NTD: N-terminal domain, RBD: Receptor-binding domain, FP: Fusion peptide, HR1, and HR2: Heptad-repeat regions 1 and 2, respectively, TM: Transmembrane region, CT: Cytoplasmic tail. c,d, Mock- and hACE2-293T cells on 96-well plates were infected with MLV PV (5 x 108 vector genome per well) expressing GFP and pseudotyped with the indicated viral glycoprotein and analyzed 24 h later. Representative histograms (c) or mean ± SEM (d) of five experiments conducted using two independent PV preparations are shown. Each dot in (d) indicates an average value of a duplicated experiment. Significant differences were analyzed by two-way ANOVA with Sidak multiple comparisons test. PV titers are presented in Extended Data Fig. 1. FKO: Furincleavage knockout mutant.
Epidemiological explanations
Researchers have been baffled by the fact the COVID-19 pandemic quickly infected a large population in countries such as Italy and cities such as New York. The reason behind this could be the increased transmissibility of the virus due to the mutations, say experts. In these regions, the health care system was overwhelmed soon, and deaths were also higher. On the other hand, in areas such as San Francisco and Washington State, the infection did not cause a rapid overpowering of the health care system. The cause could lie in the community or in the virus itself.
Conclusions and implications
The researchers wrote that “SG614 is more stable than SD614”. This corroborates with the data obtained from various parts of the world. This implies that those viruses that contain SG614 are more likely to be transmitted from one individual to another.
The viruses they engineered for this study or the pseudovirions are harmless, the researchers explained. They called for more population-based epidemiological studies to understand the nature and effects of the mutation in the viral strains that infect humans.
Farzan explained that the strains they studied were from databases such as GenBank, where the genetic data from the coronaviruses from around the world are pooled. He said that in February, the database showed that there was no D614G mutation. By March, one-fourth of the samples deposited in the database had this mutation. In May, they noted, nearly 70 percent of the samples of the viruses had this mutation that made it increasingly infectious. He said, “Over time, it has figured out how to hold on better and not fall apart until it needs to. The virus has, under selection pressure, made itself more stable.” Choe added that it is still not clear if the mutation makes the virus more deadly to those infected.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Sources:
Journal references:
- Preliminary scientific report.
The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity, Lizhou Zhang, Cody B Jackson, Huihui Mou, Amrita Ojha, Erumbi, S Rangarajan, Tina Izar, Michael Farzan, Hyeryun Choe, bioRxiv 2020.06.12.148726; doi: https://doi.org/10.1101/2020.06.12.148726
- Peer reviewed and published scientific report.
Zhang, Lizhou, Cody B. Jackson, Huihui Mou, Amrita Ojha, Haiyong Peng, Brian D. Quinlan, Erumbi S. Rangarajan, et al. 2020. “SARS-CoV-2 Spike-Protein D614G Mutation Increases Virion Spike Density and Infectivity.” Nature Communications 11 (1). https://doi.org/10.1038/s41467-020-19808-4. https://www.nature.com/articles/s41467-020-19808-4