A large-scale single-cell screening endeavor on a wide variety of animals showed that cells targeted by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are most abundant in cats, but are also found in other animals – unveiling a valuable resource for controlling the ongoing disruptive coronavirus disease (COVID-19) pandemic. The study is freely available on the bioRxiv* preprint server.
The co-evolution of viruses and hosts has been underway for millions of years, collectively shaping the immune landscape of various animal species, as well as the host range of different viral agents.
The bat has been put forward as the original host of SARS-CoV-2; nonetheless, the transmission from bats to humans necessitates intermediate hosts as well. Several studies have linked pangolins, dogs, cats, and hamsters with SARS-CoV-2 infection and subsequent transmission.
.jpg)
Novel Coronavirus SARS-CoV-2: This scanning electron microscope image shows SARS-CoV-2 (round gold objects) emerging from the surface of cells cultured in the lab. SARS-CoV-2, also known as 2019-nCoV, is the virus that causes COVID-19. The virus shown was isolated from a patient in the U.S. Credit: NIAID-RML

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Moreover, viral receptors can be considered as fundamental determinants for virus entry. Hence understanding receptor expression patterns are crucial for elucidating intra-species and inter-species transmission of viruses.
Previous studies have shown that animal tissues have a high heterogeneity in terms of cellular composition and gene expression profiles. At the same time, the ACE2 receptor (which is used by SARS-CoV-2 for cell entry) is only expressed in a small array of specific cell populations.
Consequently, the single-cell analysis of SARS-CoV-2 target cells represents a rather enticing field to investigate, and this complex research area was recently tackled by a large number of researchers from China, Denmark, the United Kingdom, and the United States.
Devising a single-cell transcriptome analysis
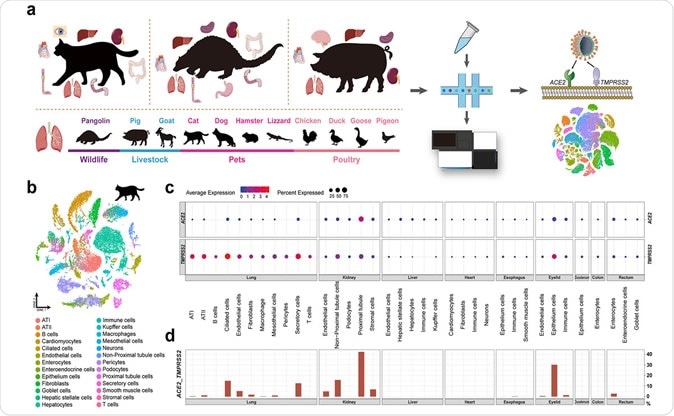
The researchers built the single-cell atlas for pets, livestock, poultry, and wildlife. They then screened putative SARS-CoV-2 target cells, indicated by the co-expression patterns of ACE2 entry receptor and TMPRSS entry activator. A systematic evaluation of their susceptibility followed this.
A total of 11 samples were collected for the purposes of this study, including four pets (cat, dog, hamster, and lizard), two livestock (pig and goat), four poultry (chicken, goose, duck, and pigeon) and one wild animal (pangolin).
The pets were bought from a pet market, while the livestock and poultry were purchased from an agricultural market. In addition, the sole pangolin sample was collected from a dead animal and immediately stored in a freezer after dissection.
"To investigate the susceptibility of host cells to different kinds of viruses, we screened the expression patterns of 114 receptors of 144 viruses (representing 26 virus families) in distinct organs of the cat, pangolin, and pig and the lung of pets, livestock, poultry and wildlife in an unbiased manner, resulting in a comprehensive atlas of virus target cells", explain study authors the crux of their methodological approach.
Cell clustering was performed with the use of a Seurat computational clustering tool, whereas cell-type annotation has been conducted in accordance with cluster differentially expressed genes and the expression of canonical cell type markers.

SARS-CoV-2 target cells predominate in cats
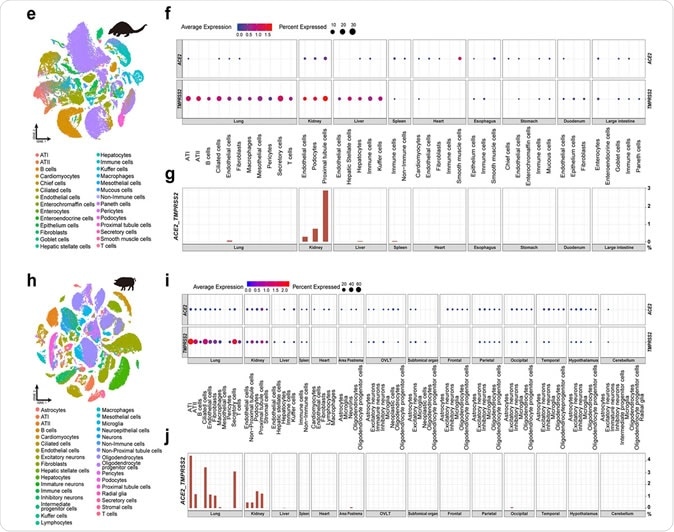
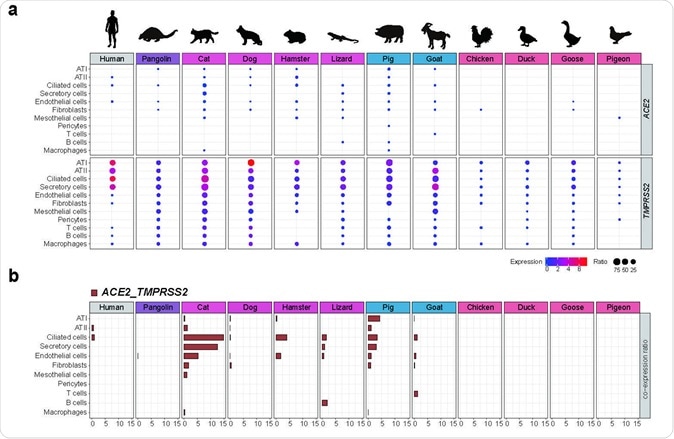
This study identified the presence of SARS-CoV-2 target cells in distinct tissues of cat, pangolin, and hamster, which was indicated by the concomitant expression of SARS-CoV-2 entry factors: ACE2 and TMPRSS2 (the latter being responsible for priming the viral spike protein used for cell entry).
The most notable finding was that the proportion of SARS-CoV-2 target cells in the cat was considerably higher than other investigated species; however, SARS-CoV-2 target cells were also detected in multiple cell types of domestic pig.
"This implies the necessity to carefully evaluate the risk of cats during the current COVID-19 pandemic and keep pigs under surveillance for the possibility of becoming intermediate hosts in future coronavirus outbreaks", elucidate study authors.
When frequencies of SARS-CoV-2 target lung cells were compared across different species, the researchers noticed that the cat clearly outweighed other species. More specifically, the percentage was 13.2% in ciliated feline cells, compared to pigs (3.4%) and hamsters (3.9%).
Furthermore, the obtained points towards the conclusion that the co-expression of ACE2 and TMPRSS2 is quite rare in dog lung cells, and completely absent in poultry lung cells (i.e., chicken, goose, duck, and pigeon).
A novel approach as a blueprint for future research
This study definitely reveals a novel strategy to discover putative susceptible hosts, as the distribution of viral target cells in distinct cell populations enables susceptibility screening of all existing viruses (by utilizing virus receptor information) on all existing species (utilizing single-cell RNA sequencing data) in an unbiased manner.
"We anticipate that the information gained from the present study will certainly augment future research work, and provide some novel insights about the prevention and control strategies against SARS-CoV-2 along with many other harmful viruses", conclude study authors.
And indeed, with the development of single-cell sequencing techniques and the unprecedented number of collaborative projects tackling this research question, the atlas for additional species could be generated at an accelerated pace. This could, in turn, help in controlling the current pandemic, but also serve as an early warning system for future threats.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Chen, D. et al. (2020). Single-cell screening of SARS-CoV-2 target cells in pets, livestock, poultry and wildlife. bioRxiv. https://doi.org/10.1101/2020.06.13.149690.
- Peer reviewed and published scientific report.
Chen, Dongsheng, Jian Sun, Jiacheng Zhu, Xiangning Ding, Tianming Lan, Xiran Wang, Weiying Wu, et al. 2021. “Single Cell Atlas for 11 Non-Model Mammals, Reptiles and Birds.” Nature Communications 12 (1). https://doi.org/10.1038/s41467-021-27162-2. https://www.nature.com/articles/s41467-021-27162-2.