The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), causing the current pandemic of COVID-19, is the focus of many investigators seeking to find an effective vaccine or therapeutic. Among the potential targets are various protease enzymes suspected of allowing the virus to gain entry to host cells and establish a successful infection.
Proteases in Human Lung Tissue
MSPL and TMPRSS13 have both been found to be part of the cDNA found in human lung tissue. Both are part of the type II transmembrane serine protease (TTSP) family, which has a transmembrane domain near the N-terminal end and a serine protease domain at the C-terminal end.
Other TTSP characteristics include a single-chain structure in the inactive form, which is activated by cleavage at a highly conserved activation motif, to become the single-chain active form. The two chains of the active enzyme are linked by a disulfide bridge, allowing the enzyme to remain bound to the membrane.
The catalytic serine protease domain has a highly conserved three-residue region, consisting of His, Asp, and Ser. The Asp is conserved, at the bottom of the substrate-binding pocket of the S1, indicating that the pocket specifically binds substrate with either P1 Arg or Lys.
Among the four subfamilies of TTSPs, these two enzymes belong to the Hepsin/TMPRSS subfamily. MSPL and TMPRSS-2 both have a low-density lipoprotein (LDL) receptor A domain towards a domain called the SRCR domain in the stem region, which has a compact β-sheet wound around an α-helix fold with 2-4 stabilizing disulfide bonds. The LDLA domain is smaller and has six conserved cysteine amino acids that help form disulfide bonds and a calcium ion, both of which stabilize its structure.
Only Hepsin has an available 3D structure in this whole family, but it does not have this LDLA domain. The SRCR domain of MPSL is rotated to 80 degrees against the corresponding domain of hepsin when both are fitted by SPD, perhaps because of the LDLA domain. In other words, the SRCR and SPD domains are compactly packed in MPSL than in hepsin, which has a more widely splayed configuration.
TMPRSS2 and MSPL in Influenza Virus Pathogenesis
A recent report described the role of TMPRSS-2 and MSPL in the spread of the high pathogenic avian influenza (HPAI) virus. These protease enzymes cleave the viral hemagglutinin (HA) antigen, a glycoprotein on the surface of the influenza virus, into two subunits, HA1 and HA2. This cleavage is necessary for the HA1-mediated binding of the virus to the host cell and the onset of endocytosis, while HA2 facilitates the fusion of the virus and endosome.
As of now, the seasonal influenza viruses have two HA processing motifs, one having a single arginine, recognized by TMPRSS2, and the other polybasic. The latter is seen in highly pathogenic avian influenza (HPAI) viruses at the cleavage site. MSPL and its splice variant TMPRSS-13 bind to both motifs. The multibasic motif is also recognized by furin and proprotein convertases that are found in most body tissues within the Golgi network.
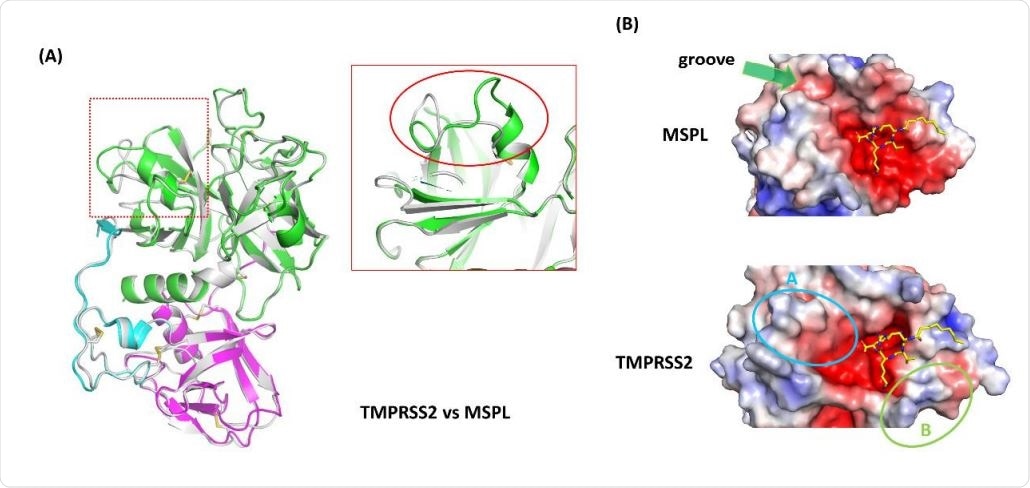
Homology model analysis of TMPRSS2. (A) A homology model of TMPRSS2 (gray ribbon) was built using MSPL as a template. Superposed analysis revealed a large structural difference at β12-β13 loop region (red rectangle). (B) Electrostatic surface potential of MSPL and TMPRSS2 SPD. MSPL has a narrow groove that fits with the downstream peptide chain (green arrow). In TMPRSS2, the groove was widened and the peptide binding site become a bowl-shaped (cyan oval A). A positively-charged area derived from Lys225 is indicated in green oval B. The potential map is colored by from red (-5kT/e) to blue (+5kT/e).
Furin Inhibitor Interaction with MSPL
Earlier, researchers found that MPSL can be inhibited using a furin inhibitor, which mimics the substrate of furin, namely, decanoyl-RVKR-cmk peptide. When activated by cleavage, and perhaps because of it, MSPL forms a salt bridge just before the catalytic site. The salt bridge may also contribute to the formation of the S1 pocket and the oxyanion hole through the resulting conformational changes.
The SPD domain of MSPL shows the same structure as the S1 serine proteases trypsin and chymotrypsin. Furin inhibitor peptides bind covalently to some catalytic residues in the SPD. The side chain of P1-Arg inserts and binds deep into the highly conserved S1 pocket, and the oxyanion hole is directly bound to the carbonyl oxygen of this residue as well.
Salt-bridges and hydrogen bonds are also formed between some residues which are in the bottom of the pocket. This pattern of interaction is characteristic of the S1 serine proteases.
On the other hand, the residues that interact with P2-Lys are not conserved in the Hepsin/TMPRSS2, which could be why most of them recognize only the single basic motif. The P2-Lys side chain interacts with the 99’s loop, which is rich in Asp and Glu and found only in MSPL.
Furin Inhibitor Interaction with Furin and MSPL
The furin inhibitor:furin combination has the inhibitor in an extended configuration, while the MSPL:furin inhibitor complex contains the inhibitor molecule showing a bend at the valine of the P3 – this difference means that both furin and MSPL can bind the inhibitor.
The investigators found that the structure of MPSL has very different arrangements of SRCR and SPD domains from hepsin, which explains how the former recognizes both motifs. Subsequently, they generated an optimized sequence for peptide inhibitors to the enzyme, with 220-fold greater inhibitory activity. The current paper describes the crystal structure of the extracellular region of the enzyme-decanoyl-RVKR-cmk peptide complex.
They also modeled a homology sequence for the TMPRSS2 protein and analyzed the preferential binding of the target sequence to either S1 or S2 subunit of the spike protein of the SARS-CoV-2. This could prove to be of great use in designing anti-influenza drugs, which can prevent the uptake of the virus into the host cell.
New Inhibitors More Potent
The distance between the P4-Arg in the furin inhibitor and the active site in the SPD domain of MSPL led the researchers to consider the possibility that the residues at P3 could be replaced by more basic residues to interact more readily with the acidic surface of the human enzyme and thus increase its potential to inhibit the latter.
Based on this, they designed four inhibitory sequences based on the furin inhibitor and tested their inhibition potency against MSPL. All were found to be 4-10-fold more potent as inhibitors of recombinant MSPL compared to decanoyl-RVKR-cmk peptide. Again, they were inhibited furin 200 times more powerfully than the original peptide.
Applications of Structural Studies
In unpublished data, an immunofluorescence assay carried out to examine the inhibitory activity of one of the new peptides reports that this was more effective than the furin inhibitor at inhibiting infection with the mutant influenza virus. This is an important finding since no therapeutic drug has yet been found against the HPAI virus.
MSPL has also been found to play a role in cleaving the spike protein and activating infection by SARS-CoV and MERS-CoV, which could help evolve drugs to treat these conditions as well.
Secondly, the structure also helps predict the tertiary structure of TMPRSS2. The homology model shows a very similar alignment of the domains between TMPRSS2 and MSPL, with the majority of disulfide bonds being conserved.
The most significant change is in the SPD domain, between the beta-12 and beta-13 loop regions. The alteration in the structure in this region produces a large groove for substrate binding, exposing the protease to the target peptide for easy capture. Another change is the substitution of Glu in MSPL by Lys in TMPRSS2, which causes the latter protease to prefer a monobasic target like alanine or valine.
The researchers say, “This homology model well reflects the feature of TMPRSS2 target peptide recognition.”
Thirdly, the researchers suggest that the calcium-binding sites in the LDLA domain could help the protease bind to basic residues like lysine or arginine via electrostatic forces when these amino acids are near the active SPD sites in the proenzyme form of MPSL. This is supported by the structure of the LDL receptor, indicating the presence of internal interactions between the calcium-binding sites of the fourth and fifth domains, and the lysine amino acid of the beta-propeller domain.
As a result of these internal interactions, ligands such as LDL cannot bind to the calcium-binding sites. However, it is not clear whether there are such interactions between the LDLA and SPD domain, without data on the structure of the proenzyme form of either MSPL or other TMPRSS variants that have these three domains. If so, the substrate may not be able to access the active site.
Some mutations do prevent the autoactivation of TMPFSS2 by inhibiting proteolytic cleavage at the SPD-SRCR interface. The deficiency of TMPRSS13 leads to abnormal skin development, but the mechanism remains unclear. MSPL and furin can cleave myostatin, which inhibits myogenesis, which in turn causes muscle atrophy. These findings, which are related to its biological function, are useful to expand knowledge in this field.