The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus has been spreading rampantly over the whole world, infecting millions of people (over 8.45 million at last count), with over 452,000 deaths, as of June 19, 2020. It causes a broad spectrum of disease manifestations, from asymptomatic disease to severe or even fatal respiratory symptoms.
Acute respiratory distress syndrome (ARDS) and multi-organ dysfunction occur more often in high-risk individuals, especially the elderly and those with cardiovascular disease, obesity, diabetes, and immunosuppression.
The infection depends on the presence of the main protease, which is required for the replication of the virus. The importance of this protease in viral replication has made it a prime target for developing drugs against the virus. It is necessary to elucidate the structure of this enzyme in order to develop or identify an effective inhibitor.
The main protease is a cysteine protease with considerable homology to the corresponding enzymes of other coronaviruses (CoV). It has a catalytic site formed by two residues, His41 and Cys145. Many studies have examined the efficacy of covalent peptides, which closely mimic the structure of the protease. Unfortunately, these properties also mean that they can create toxic effects as a result of off-target interactions with the host proteins.
What is Shikonin?
A previous investigation focused on a Chinese herbal product called shikonin. The extract itself is derived from the root of the plant Lithospermum erythrorhizon. Shikonin and its derivatives have antiviral, antibacterial, and anti-inflammatory effects, as well as tumor inhibition properties. It is a non-covalent inhibitor of the main protease of SARS-CoV-2.

Lithospermum Erythrorhizon. Image Credit: JIANG HONGYAN / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The current study aims at discovering the crystal structure of this protease-shikonin complex. The findings show that the binding occurs by a novel mechanism that could result in the development of new inhibitors that avoid covalent inhibition of the main protease.
The Overall Structure
The researchers found that the crystal structure of the main protease in the protease-shikonin complex had the same overall fold as described for a previous structure in the apostate, but with some crucial differences in the complexed state.
The conformation is very different in the apoenzyme state from that in the inhibitor-bound state. For one, the amino acids in the oxyanion hole and the N-finger regions are disordered in the apoenzyme state, but more ordered when it is bound to the inhibitor, because of the interaction between the residues between 140-145 in a different order. These residues are vital for substrate or inhibitor binding.
The catalytic site residues undergo a conformational change, causing steric hindrance following binding with the inhibitors listed earlier.
It is important to note that in the current structure, in contrast with the apoenzyme state, both of the protease protomers have clear density, and shikonin binds only to protomer A, perhaps because it has a lower affinity for this protease. Both of the protomers A and B have the same conformation, but the oxyanion loop shows small variations.
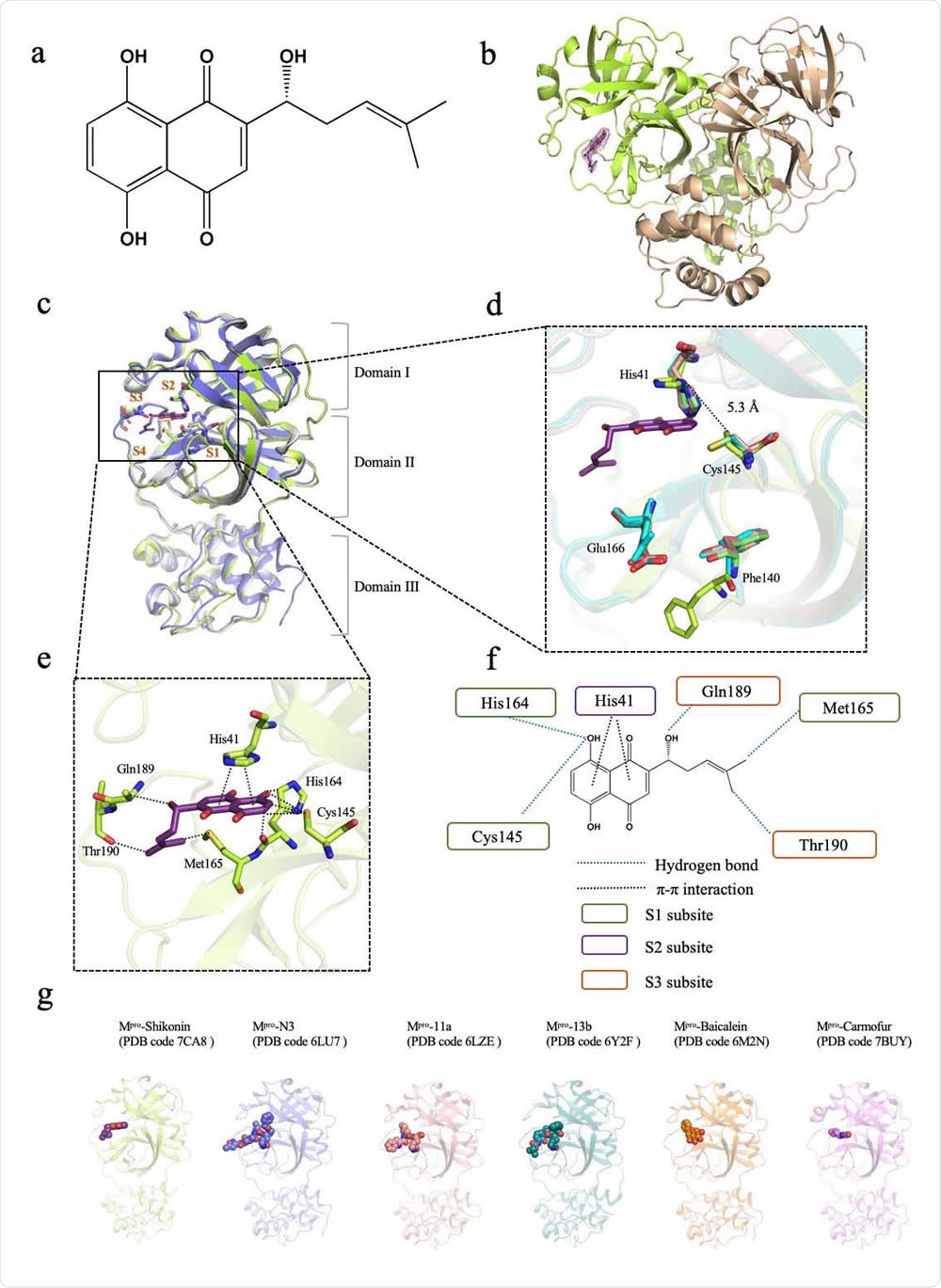
Crystal structure of SARS-CoV-2 main protease (Mpro) in complex with a Chinese herb inhibitor shikonin. a Chemical structure of the non-colavent inhibitor shikonin. b Structure of Mpro dimer. One protomer of the dimer with inhibitor shikonin is shown in green, the other is shown in wheat. The contour level is at 1σ. c Comparison of SARS-CoV-2 Mpro structures. Brown symbols S1, S2, S3 and S4 indicate the substrate binding pockets. Structure of ShiMpro is shown in green. Structure of Mpro with N3 is shown in light blue. Structure of apoMpro is shown in grey Carbon atoms of shikonin are magenta, oxygen atoms are red. Hydrogen bonds and π-π interactions are indicated by dashed black lines. d Conformational difference in catalytic site His41-Cys145. Residues of Mpro structure with shikonin are shown in green. e A zoomed view of shikonin binding pocket. f Schematic interaction between shikonin and Mpro. Hydrogen bonds and π-π stacking interactions are shown as blue dashed lines and black dashed lines, respectively. Green circle indicates conserved residues in S1 subsite. Purple circle indicates conserved residues in S2 subsite. Orange circle indicates conserved residues in S3 subsite. g Crystal structures of Mpro -inhibitor complexes from previously reported structures presenting diverse inhibitor-binding sites. Mpro structures are shown in cartoon representation and the inhibitors are shown as sphere models with transparent surfaces. The representative structures of Mpro along with covalent inhibitors, N3 (PDB code 6LU7), 11a (PDB code 6LZE) and 13b (PDB code 6Y2F) are shown. Similarly, structures for Mpro bound to natural products shikonin (PDB code 7CA8) and baicalein (PDB code 6M2N), and antineoplastic drug carmofur (PDB code 7BUY) are shown.
How Shikonin Binds to The Protease
The binding of the protease with shikonin does not affect the structure of the complex formed with previously described inhibitors. Shikonin has an inhibitor head group at the 1,4-naphthoquinone part, and a ligand tail at the six-carbon side chain. The binding pocket is a narrow cleft with four subsites around it, S1 to S4.
Shikonin has a threefold interaction with the protease at the substrate-inhibitor binding site. The S1 subsite has a triad of residues that forms hydrogen bonds with the shikonin. The aromatic head group of the shikonin interacts with His41 on S2, the imidazole group of the histamine residue flipping outward from its inward-pointing orientation, and thus allowing access to shikonin. The ligand tail has two hydrogen bonds with two S3 residues.
The difference in the shikonin-bound complex lies in the catalytic paired residues, as well as in Phe140 and Glu166. With covalently bonded inhibitors, they bind to the Cys 145 at the Sγ atom. With shikonin, the Cys 145 side-chain changes its configuration to form a hydrogen bond.
The Cys145 and the His41 have a longer distance between them than with any other main protease. The Phe140 loses its former π-π bond with His163, with the phenyl ring changing its conformation to move outward to the solvent. The Glu166 has a flexible side chain, and becomes inactive in both the apo and the shikonin-protease complexes, whereas it is ordered in the other inhibitor complexes. Since this residue is important to keep the shape of the substrate-binding pocket intact through hydrogen bonding with inhibitors that mimic the binding peptides, and with the N-terminal residue of the other protomer, it is highly conserved within the main protease family.
Water Molecules in The Active Site
The shikonin-protease complex also shows that there are two water molecules in the protease at the substrate-binding site. The first forms a series of hydrogen bonds with Phe140, His163, and Glu166 in the S1 pocket, the effect of which is to keep the oxyanion hole stable in the apoenzyme state. The other water molecule forms hydrogen bonds with His41 and Cys145 in the apoenzyme state but with shikonin in the protease-shikonin complex.
The latter does not contain either water molecule, however, which led the investigators to suggest that this model, of replacing the water molecules, could boost the inhibitory power.
Applications for Future Inhibitor Development
The study uncovers the unique mode of binding of shikonin with the main protease, via three new interactions in the substrate-inhibitor binding pocket. In contrast, the covalent peptide-mimicking inhibitors bind at the S1/S2/S4 site, while antineoplastic drug carmofur only binds to the S2 subsite and another natural product baicalein binds to the S1/S2 pocket. The protease-shikonin complex structure thus shows another mode of binding, which is non-covalent and involves striking changes in the conformation of the substrate-binding pocket.
The Chinese herbal product shikonin thus provides a promising scaffold structure that can be used to generate better antiviral drugs targeting the key main protease of SARS-CoV-2.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
La, J. et al. (2020). Crystal Structure Of SARS-Cov-2 Main Protease in Complex With A Chinese Herb Inhibitor Shikonin. bioRxiv preprint. doi: https://doi.org/10.1101/2020.06.16.155812. https://www.biorxiv.org/content/10.1101/2020.06.16.155812v1
- Peer reviewed and published scientific report.
Li, Jian, Xuelan Zhou, Yan Zhang, Fanglin Zhong, Cheng Lin, Peter J. McCormick, Feng Jiang, et al. 2020. “Crystal Structure of SARS-CoV-2 Main Protease in Complex with the Natural Product Inhibitor Shikonin Illuminates a Unique Binding Mode.” Science Bulletin, October. https://doi.org/10.1016/j.scib.2020.10.018. https://www.sciencedirect.com/science/article/abs/pii/S2095927320306885.