The current pandemic of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is not the first human coronavirus (CoV) infection to affect the world. There have been four other endemic CoVs, OC43, 229E, HKU1 and NL63, and two pandemic CoVs, SARS-CoV and MERS-CoV. However, the most intensive research efforts ever to identify the factors that contribute to the rapid transmission and high pathogenic potential of SARS-CoV-2 are underway.
.jpg)
Novel Coronavirus SARS-CoV-2: This scanning electron microscope image shows SARS-CoV-2 (round gold objects) emerging from the surface of cells cultured in the lab. SARS-CoV-2, also known as 2019-nCoV, is the virus that causes COVID-19. The virus shown was isolated from a patient in the U.S. Credit: NIAID-RML

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Viral Entry and Disease Manifestations
SARS-CoV-2 shows genetic and clinical differences with SARS-CoV; it shares around 70-80% of its genome with the 2003 virus. SARS-CoV-2 has been demonstrated to interact with Angiotensin-Converting Enzyme 2 (ACE-2), the same receptor used by SARS-CoV and CoV-NL63, to enter the host’s cells, and in particular alveolar epithelial cells. However, SARS-CoV-2 has acquired novel functions that promote its harsh disease phenotype.
SARS-CoV-2 is associated with severe lung inflammation as well as inflammatory damage of the endothelial cells in the heart, the kidneys, and intestines, which suggests a strong vascular component. The virus gains entry into the host cell by the ACE2 enzyme coupled with the furin protease that is essential for host cell membrane-virus fusion.
Following infection, the virus produces unexpectedly strong and uncontrolled inflammation and immune responses, which produces multi-organ damage and increased ACE2 expression, which enhances the infectious potential.
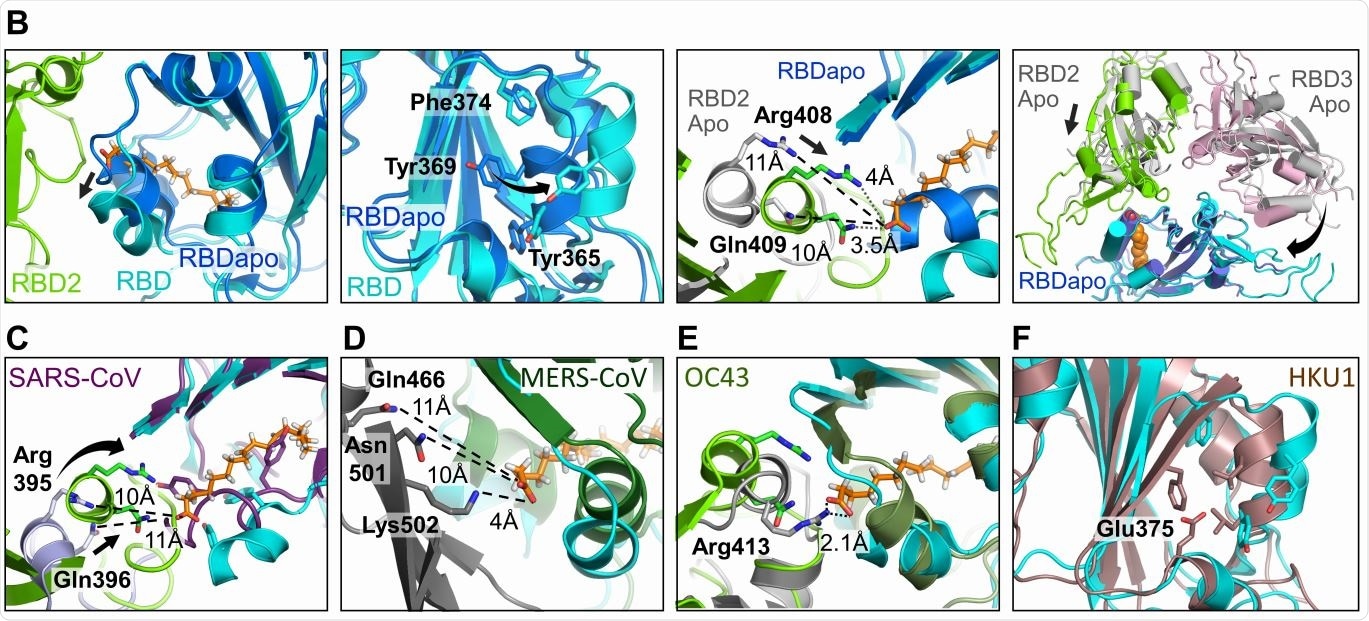
Human Coronavirus RBD architectures. (A) Alignments including the four common strains highlighting conserved residues. Residues lining the hydrophobic pocket are underlaid (cyan). Gating helix residues are marked (purple). Residues positioned to interact with the headgroup are underlaid in green. Glutamate 375 in HKU1 is underlaid in red. (B) Superimposition of RBDs in LA-bound SARS-CoV-2 (cyan, green) and ligand-free ‘apo’ form (blue, grey) (PDBID 6VXX, (6)). Gating helix movements (left two panels), approaching of adjacent RBDs upon LA-binding (center right), and compacting of the RBDs (far right) are illustrated. Rearrangements indicated by arrows. (C-E) LA-bound SARS-CoV-2 RBDs and ligand-free ‘apo’ SARS-CoV (purple, PDBID 5X58 (19)); MERS CoV (forest green, grey, PDBID 5X5F (19)) and OC43 RBDs (olive, grey PDBID 6NZK, (18)). (F) HKU1 RBD (brown, PDBID 5GNB (23)) and RBD from LA-bound SARS-CoV-2 S (cyan).
S Protein and FFA Binding
The current study looks at the structure of the Spike (S) glycoprotein using cryoEM, finding the tight binding of the protein to three molecules of linoleic acid (LA) at specific sites. This shows that the S protein has a scavenger function for LA.
Most proteins with FFA-binding pockets show the latter as a tube-like structure with a hydrophobic amino acid lining, within which the FFA’s tail fits, with a hydrophilic site that binds the FFA’s acidic head group. With the S protein, the study uncovered a hydrophobic pocket that mostly contains phenylalanine residues, forming a bent tube that fits the LA well. An Arg and Glu residue, respectively, in the receptor-binding domain (RBD) in the S protein, anchor the carboxyl group in the FFA’s head group, which is important in shaping the binding site.
The closed S protein contained LA in all three pockets, leading the scientists to conclude that the LA was bound only from the many FFAs in the culture medium. The fact that the molecule was held firmly throughout the subsequent processing steps shows the specificity and high-affinity binding.
The structure of the S protein is in the apo state before it binds to LA, but during infection, it binds to LA through a gating helix at the entrance of the binding pocket. As this binding occurs, with adjacent RBD residues locking down on the FFA head group, the trimer becomes more compact.
Features of the LA-Binding Pocket
The LA-binding pocket is also present in both the SARS-CoV and the MERS-CoV. The four essential features appear to be: conserved hydrophobic pocket, gating helix, amino acids that interact with the LA head group, and a loose apo form of the RBD. These are not all present in the other human CoVs.
MERS Coronavirus Particles Colorized scanning electron micrograph of MERS virus particles (yellow) both budding and attached to the surface of infected VERO E6 cells (blue). Image captured and color-enhanced at the NIAID Integrated Research Facility in Fort Detrick, Maryland. Credit: NIAID
The current structure does not show a direct interaction between the LA binding site and the receptor-binding motif (RBM). Still, the changes in the conformation of the RBD are possibly linked to ACE2 binding, and viral infective potential. The specific binding of the S protein to LA suggests a well-fitting pocket, which binds only LA.
A reduction in LA has been observed in COVID-19 patients, showing that a remodeling of the metabolome is occurring, as commonly observed in viral infections. When LA is supplemented, viral replication is suppressed, as also with arachidonic acid (AA) supplementation.
Lipid metabolism alterations affect three cell pathways, one related to energy homeostasis, one to biological membrane fluidity and elasticity, and the third to lipid cell signaling molecules.
Regarding membrane properties, which are thought to be altered in acute respiratory distress syndrome and severe pneumonia, typical of severe COVID-19 as well, it is important to note that disturbances in the LA pathway could change the phospholipid bilayer composition. This may well result in reduced surface tension in the lungs and impaired lung function.
When the LA pathway is remodeled to AA metabolism, cell signaling pathways change considerably because LA is the beginning point for the synthesis of eicosanoids, which are important inflammatory cytokines.
Applications of LA Binding
The researchers conclude that the LA scavenging carried out by SARS-CoV-2 can result in a non-tissue-specific disruption of normal regulation of immune and inflammatory pathways. This could be how the current virus produces such harsh features of the disease.
LA binding to the S protein causes the hydrophilic anchor to switch to the closed-form. This could, therefore, be a target for the development of small-molecule drugs, which lock S in the closed configuration irreversibly, inhibiting viral infection.
The study sums up, “Our findings provide a direct structural link between LA, COVID-19 pathology and the virus itself and suggest that both the LA-binding pocket within the S protein and the multi-nodal LA signaling axis, represent excellent therapeutic intervention points against SARS-CoV-2 infections, particularly in patient groups with increased risk due to metabolic preconditions.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Toelzer, C. et al. (2020). Unexpected free fatty acid binding pocket in the cryo-EM structure of SARSCoV-2 spike protein. bioRxiv preprint. doi: https://doi.org/10.1101/2020.06.18.158584. https://www.biorxiv.org/content/10.1101/2020.06.18.158584v1
- Peer reviewed and published scientific report.
Toelzer, Christine, Kapil Gupta, Sathish K. N. Yadav, Ufuk Borucu, Andrew D. Davidson, Maia Kavanagh Williamson, Deborah K. Shoemark, et al. 2020. “Free Fatty Acid Binding Pocket in the Locked Structure of SARS-CoV-2 Spike Protein.” Science, September. https://doi.org/10.1126/science.abd3255. https://www.science.org/doi/10.1126/science.abd3255.