Researchers from Germany and Switzerland revealed functional effects of individual severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) proteins on cellular level after infection. They placed the findings into the context of host signaling pathways in order to identify vulnerable parts of the virus amenable to treatment. The study is currently available on the bioRxiv* preprint server.
Coronaviruses penetrating in human cell. Binding of the coronavirus spike protein(red) to an ACE2 receptor (blue) leads to the penetration of the virus in the cell. 3d rendering. Image Credit: Juan Gaertner / Shutterstock
The sudden global emergence of SARS-CoV-2, the causative agent of an ongoing coronavirus disease (COVID-19) pandemic, prompted the need for an in-depth apprehension of molecular functions of viral proteins and their respective interactions with the host proteome.
Several omics studies have broadened our knowledge of COVID-19 pathophysiology, including some that were focused on proteomic aspects. For example, a comprehensive SARS-CoV-2 human protein-protein interaction map (derived with affinity-purification mass spectrometry) was recently published in the leading scientific journal Nature.
But albeit multi-omics studies (together with structural biology methods) are anticipated to have a fundamental role in the efficient clinical management of this pandemic, systems-level molecular studies of SARS-CoV-2 are hampered by the tremendously contagious nature of the pathogen.
However, there are scientists ready to tackle such challenging research questions. In order to understand the exact way how SARS-CoV-2 and related coronaviruses manipulate the host, a research group from Germany and Switzerland recently characterized proteome, interactome, and signaling processes in a systems-wide manner.
The integrative -omics approach
These researchers investigated closely related SARS-CoV-2 and SARS-CoV viruses (the latter being the causative agent of the original SARS outbreak), as well as the influence of SARS-CoV-2 on the proteome, transcriptome, ubiquitinome, and phosphoproteome of a lung-derived human cell line.
More specifically, to acquire information on the concerted activity of viral proteins during the infection, they have infected A549 cells expressing ACE2 receptor with SARS-CoV-2 and characterized the impact of viral infection on RNA transcription, protein abundance, ubiquitination (i.e., bonding of a ubiquitin-protein to a substrate protein) and phosphorylation in a time-resolved manner
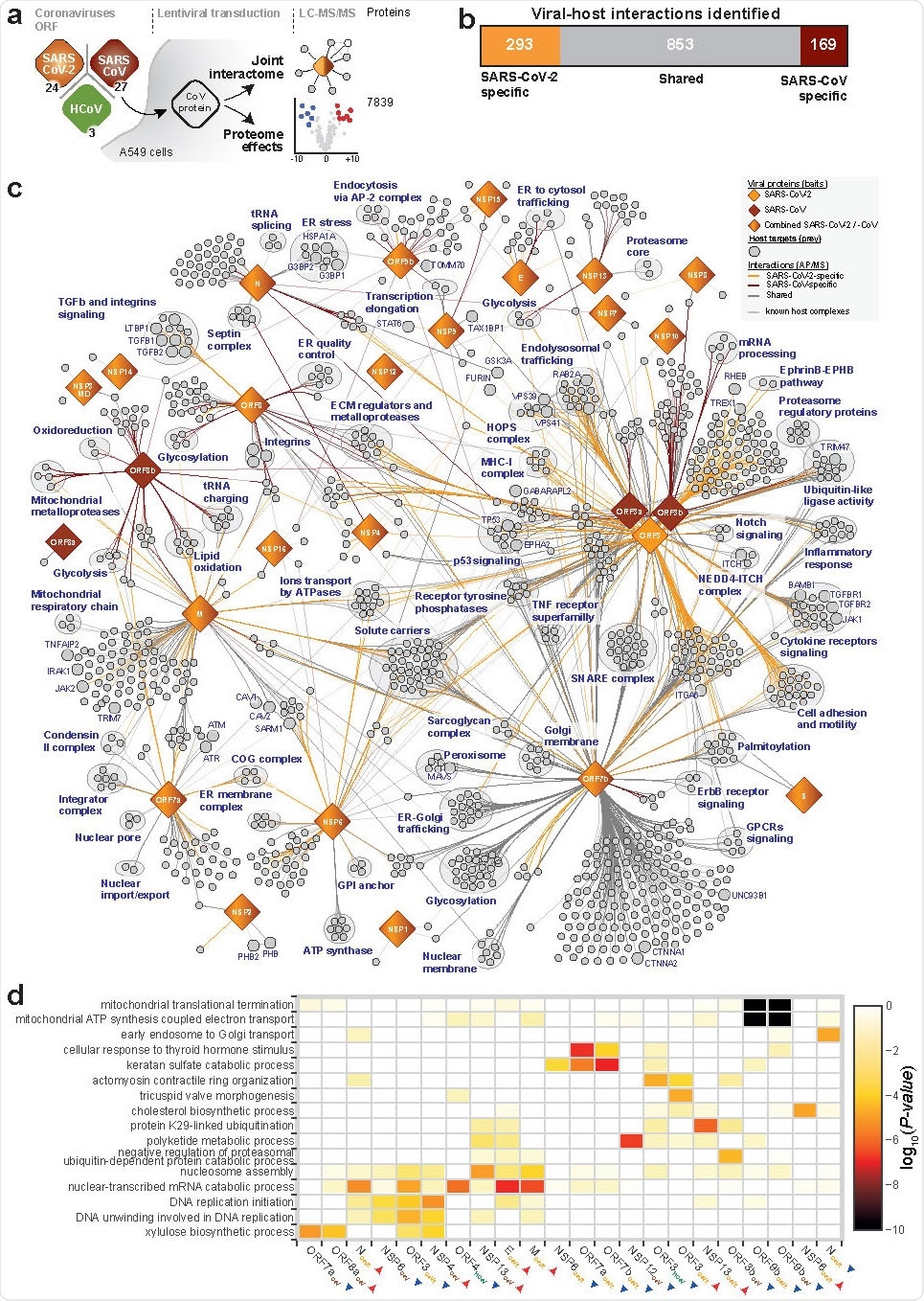
"For each viral protein, we mapped the collected data onto the global network of cellular interactions and applied a network diffusion approach," further explain study authors. "Such analysis identifies short links of known protein-protein interactions, signaling and regulation events that connect the interactors of the viral protein with the proteins affected by its expression," they add.
Such an analysis of proteome changes (induced by each viral protein) and consideration of subsequent protein interactions provided direct insights into their functions. Finally, the researchers tested a panel of 48 drugs modifying the pathways perturbed by the virus for their effects on SARS-CoV-2 replication.
Host-perturbation strategies of SARS-CoV-2
The virus-host interaction network that stemmed from this comprehensive research endeavor revealed a myriad of cellular activities intercepted by both SARS-CoV-2 and SARS-CoV. The systematic proteome and interactome profiling of individual viral proteins opened the door for a deeper understanding of their molecular mechanisms.
More specifically, when SARS-CoV-2 is concerned, this study found that it can interact with specific protein complexes, contributing in turn to a range of biological processes. Moreover, the virus can also target stress response components and DNA damage response mediators.
"Upon virus infection, we observed upregulation of TGF-β and EGFR pathways, which modulate cell survival, motility, and innate immune responses," said study authors. "Besides promoting virus replication, activation of these pathways has been implicated in fibrosis, one of the hallmarks of COVID-19", they further emphasize.
According to the drug testing results, B-RAF, JAK1/2, and MAPK inhibitors led to a significant increase of viral growth in the laboratory infection setting; conversely, tyrosine kinase inhibitors, the mTOR inhibitor, or inducers of DNA damage led to the suppression of SARS-CoV-2.
In a nutshell, projecting the obtained data onto the global network of cellular interactions unveiled relationships between the perturbations that arise upon SARS-CoV-2 infection at different layers, but also identified unique and characteristic molecular mechanisms of SARS coronaviruses.
Streamlining the quest for targeted antiviral compounds
Taken together, the illustrated viral-host protein-protein interactions and pathway regulations that were observed at multiple levels recognize potential areas of SARS-CoV-2 that could be targeted by well-characterized selective antiviral drugs.
"The results highlight the functionality of individual proteins, as well as vulnerability hotspots of SARS-CoV-2, which we targeted with clinically approved drugs", explain study authors. "We exemplify this by identification of kinase inhibitors, as well as matrix metallopeptidase inhibitors with significant antiviral effects against SARS-CoV-2", they add.
Furthermore, the drug screen that was pursued in this study demonstrated the value of this type of combined dataset, and further exploration of the interplay between different -omics levels may advance the knowledge of coronavirus biology and pathogenicity.
In conclusion, these results can serve as a base for the intelligent design of combination therapies that target the virus from multiple and synergistic angles, potentiating, in turn, the effect of individual treatments, and minimizing at the same time the side-effects on healthy tissues.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Source:
Journal references:
- Preliminary scientific report.
Stukalov, A. et al. (2020). Multi-level proteomics reveals host-perturbation strategies of SARS-CoV-2 and SARS-CoV. bioRxiv. https://doi.org/10.1101/2020.06.17.156455.
- Peer reviewed and published scientific report.
Stukalov, Alexey, Virginie Girault, Vincent Grass, Ozge Karayel, Valter Bergant, Christian Urban, Darya A. Haas, et al. 2021. “Multilevel Proteomics Reveals Host Perturbations by SARS-CoV-2 and SARS-CoV.” Nature, April. https://doi.org/10.1038/s41586-021-03493-4. https://www.nature.com/articles/s41586-021-03493-4.