With the worldwide impact of the COVID-19 disease, it has become a top scientific priority to understand the molecular mechanism driving this infection. Accordingly, a mountain of sequenced genomes and transcriptomes is now publicly available.
Novel Coronavirus SARS-CoV-2 Spike Protein 3D print of a spike protein on the surface of SARS-CoV-2—also known as 2019-nCoV, the virus that causes COVID-19. Spike proteins cover the surface of SARS-CoV-2 and enable the virus to enter and infect human cells. For more information, visit the NIH 3D Print Exchange at 3dprint.nih.gov. Credit: NIH

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The Impact of COVID-19
COVID-19 is primarily known for its respiratory manifestations. The pandemic has shut down economic activity significantly in a majority of countries in the year’s first quarter, caused over 9.58 million infections and 488,000 deaths. Healthcare systems were overwhelmed in both developing and developed countries. The burden of severe illness and death has been borne disproportionately by Black and Hispanic minorities, older adults, and those with a coexisting illness such as hypertension, lung disease, and type 2 diabetes.
Public Data on Respiratory Virus Infections
The world has had several outbreaks of infection believed or proved to have jumped from animal species to humans. These include the H1N1, bird flu, SARS, and MERS epidemics. In order to understand more about how these viruses affect the host cells, high-throughput RNA sequencing, and microarray technologies help to detect and uncover the molecular changes following respiratory virus infections.
However, bioinformatic analyses of the vast amount of data available is required to extract useful information.
The Study: Finding Genes Exclusively Altered in COVID-19
The current study aimed to carry out a meta-analysis of such data to understand how SARS-CoV-2 affects the cell at the molecular level, how it is similar to and different from other respiratory viruses, and which parts of the virus-host cell interaction are susceptible to drug inhibition.
The results show that SARS-CoV-2 infected human bronchial epithelial cells show altered gene expression, some of which related to type I interferon signaling pathways, and others to the inflammatory response, immune pathways, antiviral responses and other reactions to the presence of the virus.
Secondly, the researchers analyzed five studies on human lung epithelial cells infected by other viruses and found that this also showed differential expression of various genes following infection. These differentially expressed genes (DEGs) were then compared with those found in SARS-CoV-2 infection, to arrive at 15 genes that were differentially expressed specifically in the latter. Some of these were upregulated, while others were suppressed.
A Few Genes Described
Among these are genes like the colony-stimulating factor 2 (CSF2) that encodes a key cytokine elevated in many respiratory diseases such as pulmonary alveolar proteinosis. Others include the S100 calcium-binding protein A9 (S100A9) and S100A8, which bind calcium and zinc and are found at high levels in lung inflammation.
The transcriptional and immune response regulator (TCIM) is a positive regulator of cell proliferation pathways in thyroid cancer. At the same time, the cysteine-rich C-terminal 1 (CRCT1) gene is found to act as a tumor suppressor. Meanwhile, the tripartite motif family-like 2 (TRIML2) gene enhances the growth of human mouth cancers.
Another similar gene is the C-X-C Motif chemokine ligand 14 (CXCL14), which suppresses the growth of cancer cells in the mouth, lung, and the head and neck, besides prostate and breast cancers.
Some other genes include Hephaestin-like 1 (HEPHL1), which is a glycoprotein that has ferroxidase activity, and MAS-related GPR family member X3 (MRGPRX3) that is part of the G-protein coupled receptor family. The latter is found to be reduced in expression in human bronchial epithelium after exposure to smoke from e-cigarettes.
In addition, the expression of the apoptosis pathway and type I interferon signaling pathway genes were found to be commonly affected by both SARS-CoV-2 and other viral infections.
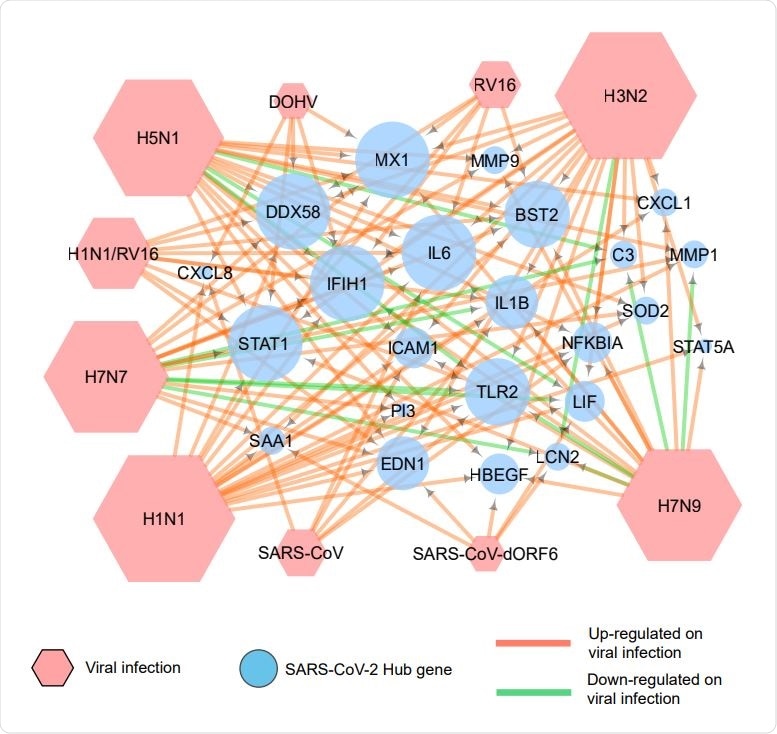
Association of COVID 19 hub genes with other respiratory viral infections. A network generated by use of Cytoscape depicts the expression of COVID-19 hub genes on infection by various respiratory viruses. The green arrows indicate down-regulation, and the red arrows show up-regulation of these genes.
CSF2: Central to SARS-CoV-2 Infection?
An analysis of over 1,000 interactions between almost 170 host cell and viral proteins shows that 24 genes may be considered as hub genes.
Among these, the only one that is exclusively expressed in cells infected by SARS-CoV-2 is the CSF2. In contrast, the others are almost always altered in cells infected by other respiratory viruses as well. The CSF2 gene is therefore suspected of having a central role in facilitating SARS-CoV-2 infection of the human lung epithelial cells.
The study concludes, “Both PPI analysis and comparative transcriptome analysis point to a role of CSF2 in the molecular mechanism of SARS-CoV-2 infections of human lung epithelium. The current study also highlights the exclusivity of known lung inflammation disorder genes such as S100A8 and S100A9 with respect to SARS-CoV-2 infection.”
These findings will now need to be experimentally validated to find optimal molecular targets that can be used to develop therapies against the virus.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources