Antiviral drug screen of kinase inhibitors, which was done by researchers from the University of California and Cedars-Sinai Medical Center, identified cellular signaling pathways critical for SARS-CoV-2 replication and revealed an important mechanism of host-pathogen interaction vital for further drug research. Their results are available on the bioRxiv* preprint server.
.jpg)
Novel Coronavirus SARS-CoV-2 Colorized scanning electron micrograph of a cell (green) infected with SARS-COV-2 virus particles (purple), isolated from a patient sample. Image captured at the NIAID Integrated Research Facility (IRF) in Fort Detrick, Maryland. Credit: NIAID
The spread of a highly contagious severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease (COVID-19), has resulted in the ongoing global health crisis with over 500,000 deaths and ten million confirmed cases to date.
However, as this is a novel virus, our knowledge of the mechanisms of COVID-19 pathogenesis is very limited. As a result, many attempts to develop targeted antiviral strategies were hampered, and the effective therapy for this unmet medical emergency is still elusive.
What we do know is that a myriad of RNA viruses can prompt significant DNA damage – even in instances where viral replication takes place solely in the cytoplasm. Subsequent activation of DNA damage response pathways can additionally boost the replication of viral RNA genomes.
Furthermore, DNA damage can contribute to the pathogenesis of RNA viruses by triggering apoptosis (i.e., programmed cell death), stimulating an inflammatory immune response, and introducing deleterious mutations that, in turn, increase the risk of cancer formation.
The objectives of this new study by US researchers were to elucidate host-pathogen interactions by defining cellular signaling pathways essential for SARS-CoV-2 replication, and also to unveil small molecule kinase inhibitors with anti-SARS-CoV-2 activity.
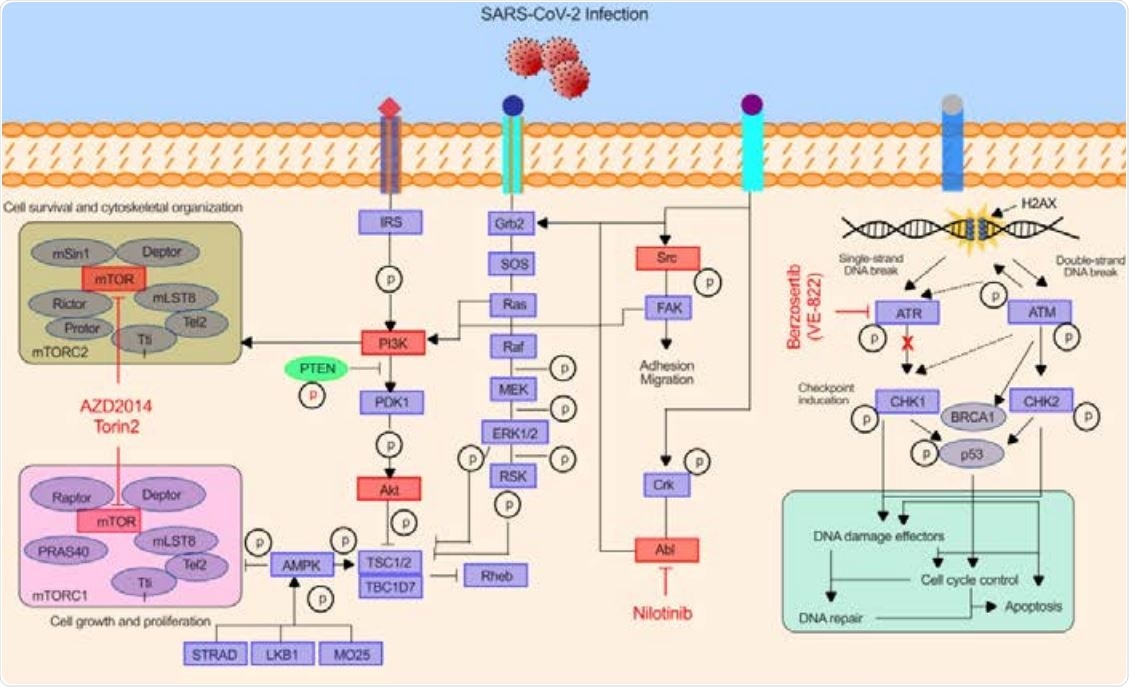
Schematic illustration 16 demonstrates the key pathways identified in the kinase inhibitor drug screen having critical role in 17 SARS-CoV-2 infection. Drug compounds and their respective target kinase pathways (mTOR18 PI3K-AKT, ABL-BCR/MAPK, and DNA-Damage Response) are described.
Devising a high-throughput drug screening system
In a nutshell, the researchers developed a high-throughput drug screening system to pinpoint antiviral drugs that could be used in the treatment of COVID-19. For that purpose, they have established a SARS-CoV-2 infectious cell culture system and virological assays by utilizing Vero-E6 cells (i.e., a cell line derived from an African green monkey), but also certain human cell lines.
This study made use of a small molecule library of 430 protein kinase inhibitors, which are currently in different stages of ongoing clinical trials. Moreover, the screening concentration with optimal activity and minimal toxicity has been precisely identified.
Most of the tested kinase antagonists in this study belong to ATP-competitive inhibitors, a class of nucleoside analogs with potent antiviral activity. They are known to halt phosphotransferase activity of their targets, but they also function beyond just blocking catalytic activity.
Cellular signaling pathways and host-pathogen interactions
From the primary screen, the researchers have identified a total of 34 compounds capable of inhibiting the viral cytopathic effect in epithelial cells. A nexus of drug and protein interactions showed that these compounds specifically target a limited number of cellular kinases.
The most important discovery is the precise identification of mTOR-PI3K-AKT, ABL-BCR/MAPK, and DNA damage response pathways as cellular signaling pathways that are indispensable for SARS-CoV-2 infection.
Subsequently, a secondary screen confirmed that compounds such as berzosertib (VE-822), vistusertib (AZD2014), and nilotinib have significant anti-SARS-CoV-2 activity, which is achieved by inhibiting key cellular enzymes that play a role in viral replication.
The researchers have also demonstrated that berzosertib, a kinase inhibitor in the DNA damage response pathway which limits viral genomic RNA replication, exhibited potent antiviral activity in a human epithelial cell line, but also in human-induced pluripotent stem cell (hIPSC)-derived cardiomyocytes.
Repurposing kinase inhibitors for COVID-19 treatment
"Overall, this present study illustrates key signaling proteins involved in SARS-CoV 2 replication and provides potential avenues for novel antiviral drug development", study authors explain the significance of their findings.
As some of the aforementioned inhibitors significantly influence the DNA damage response pathway, it appears that the latter is a valid target for host-directed therapeutic development for COVID-19; therefore, further investigation is warranted.
As of mid-June 2020, eight clinical studies based on berzosertib (phases one and two) have been underway, and the drug appears to be well tolerated with a rather mild side effect profile. In the meantime, further evaluation of the targets using CRISPR-based knockout studies is currently in progress.
Hence, the authors believe berzosertib can be rapidly repurposed to treat COVID-19 patients, following obligatory additional in vivo efficacy and safety studies. Finally, other drugs appraised in this study may show promise for clinical use in the near future.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources