
Winter moth, Operophtera brumata. Image Credit: Henri Koskinen / Shutterstock
The molecular structure has been mapped in great detail for SARS-CoV, a causative agent of the 2002-2004 SARS outbreak. It was then discovered that the mobile genetic element s2m (found in several families of single-stranded RNA viruses) has the propensity to move between distantly related viruses.
Albeit evolutionary and functional origin of s2m is still unknown, the high level of conservation detected in even distantly related viruses – notwithstanding high overall mutation rates – indicates that s2m is under strong selection.
The presence of s2m in the SARS-CoV-2 genome and other members of this group is most likely due to a single horizontal transfer event, predating the separation of the SARS-related viruses. But without more in-depth insight into its sequence, it is quite cumbersome to reach steadfast conclusions.
In this study, Dr. Torstein Tengs from the Norwegian Institute of Public Health, Professor Charles F. Delwiche from the University of Maryland in the US, and Dr. Christine Monceyron Jonassen from the Østfold Hospital Trust in Norway, aimed to characterize the specific genotype of s2m found in SARS-CoV-2.
Exploring nucleotide databases
The Basic Local Alignment Search Tool, complemented with a nucleotide query (BLASTN), was employed to search through regions of local similarity and compare various biological sequences in the entire virus section of GenBank. More specifically, all s2m sequence genotypes that were reported in the literature were utilized as query sequences.
Moreover, when s2m motifs were interrogated in insects, both the transcriptome shotgun assembly (TSA) and the whole genome shotgun contigs (WGS) databases were mined by utilizing the same approach.
For the subsequent phylogenetic analyses, sequences were precisely aligned with the use of the Clustal W algorithm (which aligns any number of homologous nucleotide or protein sequences), while the maximum likelihood analysis was performed by using integrated MEGA X tool.
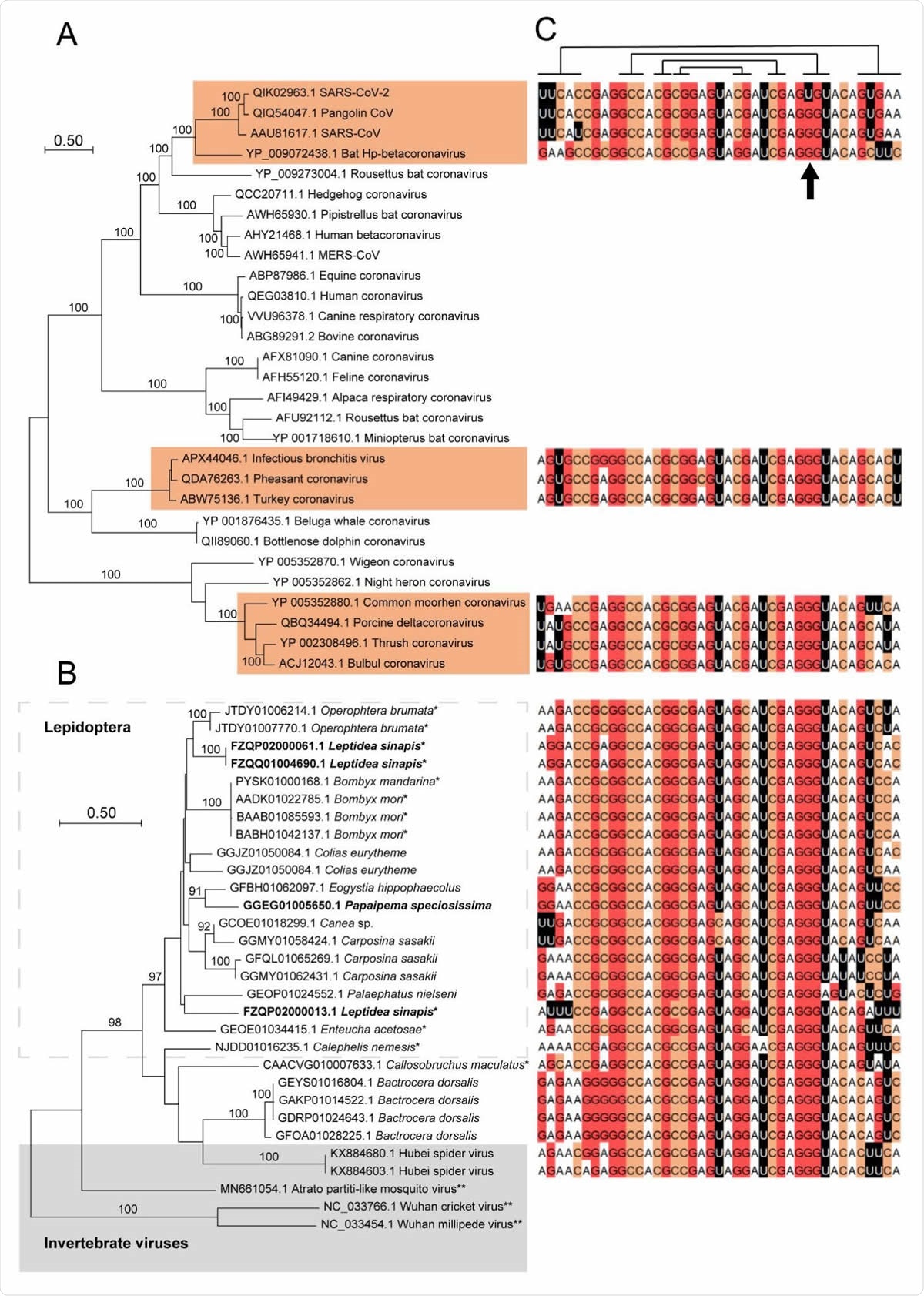
s2m in insects, arachnid viruses and coronaviruses. A) A maximum likelihood analysis was performed on ORF1ab polyprotein sequences from selected coronavirus species. s2m-containing accessions have been highlighted and bootstrap values > 90 % indicated (100 psedoreplicates). B) Maximum likelihood analysis using data from the s2m-associated hypothetical protein (see main text for details). Sequences in boldface stem from reading frames with (multiple) internal stop codons. * - genomic data, ** - accessions without s2m. C) s2m sequences corresponding to operational taxonomic units in the phylogenetic trees. Lines above alignment show (non-canonical) base-pairing residues (Robertson, et al. 2005) and the position with the unique G > U mutation in SARS-CoV-2 has been indicated.
The s2m sequence essential for viral function
The s2m genotype found in SARS-CoV-2 contains a transversion mutation (i.e., the event when a single two-ring purine is substituted with one-ring pyrimidine, or vice-versa) in position 31. More specifically, there was a notable substitution of uracil with guanine, which was consistent in all available SARS-CoV-2 accessions.
Furthermore, it was observed that this guanine is perfectly conserved outside of the SARS-CoV-2 sequences, as 100 percent of other s2m genotypes had guanine in the same position. More specifically, insects (such as long-jawed orb-weaver spiders and winter moths) were found to harbor the same genetic signature.
The insect species that contained s2m in this study were distantly related – indicating either a deep evolutionary origin with multiple losses or that the aforementioned genetic construct is also a mobile element, perhaps exploiting viruses as a vector. That is where coronaviruses come into the picture.
"Although s2m is not universally present among coronaviruses and appears to undergo horizontal transfer, the high sequence conservation and universal presence of s2m among isolates of SARS-CoV-2 indicate that, when present, the element is essential for viral function", explain study authors.
However, the exact evolutionary link between the xenologs (derived from lineage fusion or horizontal transfer) of s2m found in insects and viruses can not be ascertained based solely on the data from this study.
Mobile genetic element as a potential treatment target
"We believe that the most likely mode of transfer for s2m in viruses is through non-homologous recombination between RNA molecules", study authors summarize their findings in bioRxiv paper.
Outside the astroviruses that cause diarrheal illness, SARS-CoV and SARS-CoV-2 are the only known examples of s2m-carrying viruses that infect humans; however, it seems likely that s2m is still evolutionary active and that it will persist in affecting the evolution of positive-sense single-stranded RNA viruses (such as SARS-CoV-2).
And since the clade of s2m-containing coronaviruses (which includes both SARS-CoV and SARS-CoV-2) also includes viruses isolated from bats and pangolins, it is highly unlikely that s2m played a direct role when a zoonotic transfer is concerned.
Nevertheless, its high degree of sequence conservation hints that it may be used as a valuable treatment target. At the same time, the existence of closely related systems in insect hosts enables the fundamental biological appraisal of this apparent mobile element in somewhat tractable experimental systems.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources