A research group from the University of California Santa Barbara recently demonstrated a cost-effective, simple, and much less toxic method to isolate host and pathogen nucleic acids and proteins in order to streamline the detection of DNA and RNA viruses – including the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Their findings are currently available on the bioRxiv* preprint server.
The pressing health crisis of the ongoing coronavirus disease (COVID-19) pandemic, caused by SARS-CoV-2, has resulted not only in significant global morbidity and mortality but also in severe economic and societal consequences.
The primary weapon in our fight against this pandemic is a widespread and attainable testing approach to monitor SARS-CoV-2 burden and spread, which is needed to subsequently inform adequate containment and mitigation measures.
However, globally scaled testing is still a burning, unmet public health need; in addition, many attempts to satisfy this demand have led to shortages of supplies and reagents that are necessary for sample processing, RNA extraction, and, in turn, rapidly obtaining test results.
In other words, the extraction kits became a limiting resource for SARS-CoV-2 testing, which prompted the rise of alternative SARS-CoV-2 RNA isolation techniques and protocols that circumvent entirely the RNA extraction step.
To further address this issue, the researchers from the University of California Santa Barbara recently developed a rather simple method to isolate nucleic acids and proteins from cells and viruses known as PEARL (i.e., an acronym for Precipitation Enhanced Analyte RetrievaL).
How PEARL works and how was it benchmarked?
In a nutshell, PEARL utilizes a non-ionic detergent-based lysis solution for disrupting cell membranes and viral envelopes, providing at the same time conditions amenable for alcohol-based precipitation and retrieval of DNA, RNA, and proteins.
Therefore, PEARL can be viewed as a low-cost and column-free approach to isolate nucleic acids and proteins by using only common laboratory reagents. But simplified methods have to provide similar reliability in order to be accepted universally.
To benchmark their proposed method, the researchers have extracted RNA from de-identified samples positive for SARS-CoV-2 by using both newly develop PEARL approach and a standardly used, dedicated RNA extraction kit.
Their next step was to utilize the isolated RNA to study the levels of the SARS-CoV-2 nucleoprotein gene, as well as the host ribonuclease P genetic material in the samples. The one-step reverse transcription-quantitative polymerase chain reaction reference test for COVID-19 (recommended by the US CDC) was then applied.
After testing different samples to PEARL-lysis-solution ratios, the researchers have observed that PEARL required a rather modest increase in the initial sample input (1.25-fold) to attain sensitivity akin to the commercial RNA extraction kit used in this study.
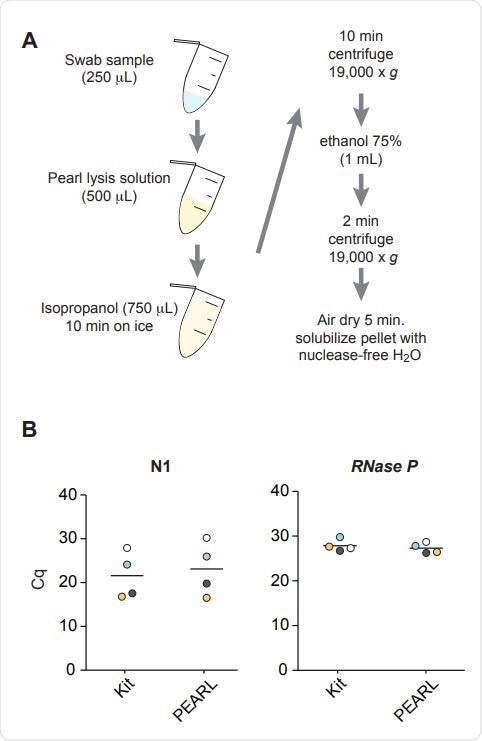
Overview of PEARL. B) Comparative RT-qPCR analysis of the levels of SARS-CoV-2 nucleocapsid (N1) and RNaseP RNA sequences in four de-identified SARS-CoV-2 positive samples after RNA extraction using PEARL or a dedicated RNA extraction kit (QIAamp Mini Elute Virus Spin Kit). Corresponding samples are color-coded. The black lines indicate the median. P = 0.72 (N1), P = 0.56 (RNaseP), t-test.
A scalable, rapid and reliable technique
"Our results indicate that PEARL facilitates the detection of SARS-CoV-2 transcripts in COVID-19 positive swab samples with sensitivity comparable to that afforded by commercially available RNA extraction kits", say study authors in their bioRxiv paper.
This particular outcome emphasizes the plausibility of using PEARL as a feasible alternative to facilitate and advance the sensitive detection of SARS-CoV-2 in respiratory samples of patients with COVID-19.
One additional advantage of PEARL in comparison to column-based commercially available RNA extraction methods is the possibility of recovering proteins alongside nucleic acids. Naturally, this broadens the potential applicability of this powerful tool for detecting many diverse viruses.
As a consequence, neither specialized equipment nor highly trained personnel is warranted, while minimal handling requirements make PEARL very scalable – which is indeed desirable for high-volume testing operations direly needed in this pandemic.
Potential roadblocks on route to the mainstream implementation
Although the research to back these claims is solid, certain caveats have to be taken into account. For example, there is a possibility that the collection medium used for sample storage prior to processing may substantially impact the performance of PEARL.
Furthermore, the extraction bias introduced by PEARL cannot be excluded, as short RNAs – including transport, small nucleolar, and micro RNAs – are much more cumbersome to precipitate in comparison to longer DNA and RNA molecules.
Hence, additional improvements may be necessary to fully implement PEARL as a mainstream nucleic acid and protein isolation tool to detect other viruses from other sources (since the sample type may significantly influence the overall performance).
But taking everything into account, it is quite clear that PEARL can offer an accessible alternative for diagnostic streamlining in geographic locations that lack professional laboratory capacity and even access to specialized reagents.
This means that the high bar of "luxury testing" can finally be lowered in many different parts of the world. And nothing is needed more in the times when COVID-19 pandemic still seems like an uphill battle.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Ponce-Rojas, J.C. et al. (2020). A Fast and Accessible Method for the Isolation of RNA, DNA, and Protein to Facilitate the Detection of SARS-CoV-2. bioRxiv. https://doi.org/10.1101/2020.06.29.178384.
- Peer reviewed and published scientific report.
Ponce-Rojas, Jose Carlos, Michael S. Costello, Duncan A. Proctor, Kenneth S. Kosik, Maxwell Z. Wilson, Carolina Arias, and Diego Acosta-Alvear. 2020. “A Fast and Accessible Method for the Isolation of RNA, DNA, and Protein to Facilitate the Detection of SARS-CoV-2.” Journal of Clinical Microbiology, December. https://doi.org/10.1128/jcm.02403-20. https://journals.asm.org/doi/10.1128/JCM.02403-20.