A large group of researchers from China recently characterized microbial co-infections in the respiratory tract of hospitalized coronavirus disease (COVID-19) patients and demonstrated a high prevalence of both viral and bacterial co-infections – particularly in individuals presenting with severe disease. Their findings are currently available on the medRxiv* preprint server and may serve to improve empirical treatment regimens.
The ongoing COVID-19 pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), poses a severe threat to both public health and the global economy. At the beginning of July 2020, there were more than 11 million confirmed cases and more than 500 thousand deaths worldwide.
Novel Coronavirus SARS-CoV-2 Colorized scanning electron micrograph of a cell heavily infected with SARS-CoV-2 virus particles (yellow), isolated from a patient sample. Image captured at the NIAID Integrated Research Facility (IRF) in Fort Detrick, Maryland. Credit: NIAID

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
COVID-19 primarily affects the respiratory tract and causes viral pneumonia that may progress to acute respiratory distress syndrome. However, the infection is frequently accompanied by gastrointestinal symptoms such as abdominal pain, diarrhea, or vomiting. Recently, it was proven that SARS-CoV-2 genuinely infects the human gastrointestinal tract.
But the question remains whether SARS-CoV-2 is solely responsible for all severity stages and manifestations of COVID-19 or the microbial co-infections can have a certain effect on clinical outcome of SARS-CoV-2-infected individuals.
Consequently, by simultaneously applying cultures, colorimetric assays, and metatranscriptomic sequencing, a large group of Chinese researchers evaluated microbial co-infections in a cohort of 23 hospitalized COVID-19 patients from Guangdong (a province in the southernmost part of China).
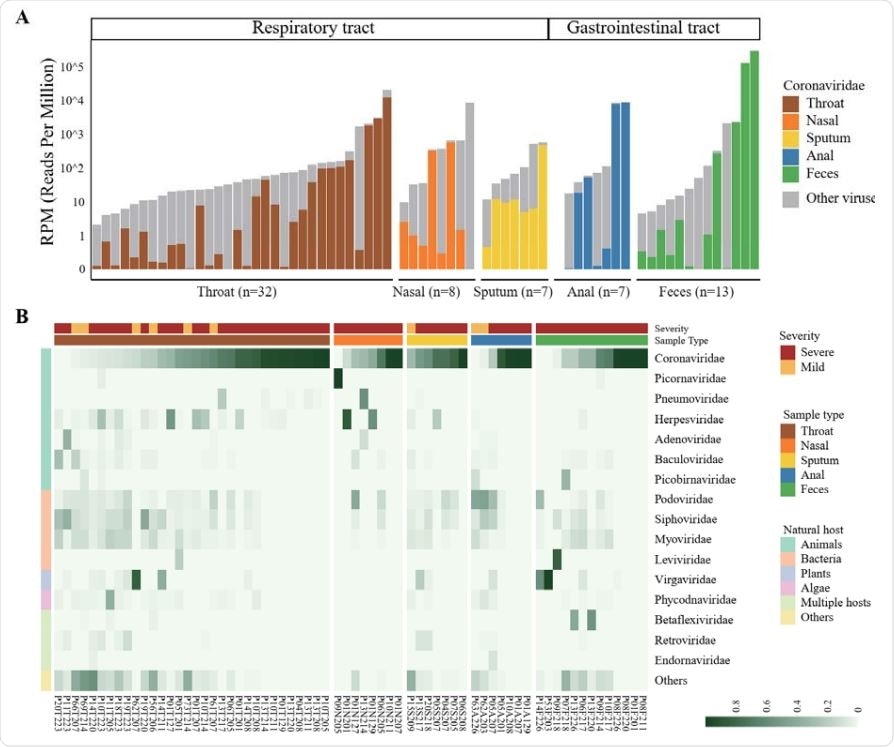
Viral RNA profiles in clinical specimens of hospitalized patients with COVID-19
From classical culture-based methods to the state-of-the-art technology
In 2020, between January 27 and February 26 (not long after the outbreak started), serial clinical specimens – primarily throat and nasal swabs, sputum and anal swabs – were collected from 8 mildly and 15 severely ill, hospitalized COVID-19 patients. All of them necessitated mechanical ventilation and were admitted to the intensive care unit.
After extracting total ribonucleic acid (RNA), ultra-deep metatranscriptomic sequencing was performed in combination with different laboratory diagnostic assays. The abundance of diverse microbial communities and co-infection rates were determined in the above-mentioned cohort of COVID-19 patients.
Prior to identifying virome and microbiota specificities, specialized software was employed to filter microbial ribosomal RNA from non-human metatranscriptomic data. Furthermore, for comparing the expression levels between different genes, normalization of target gene expression levels was performed among all detected virulence genes.
Interactions between the host, SARS-CoV-2 and microbiota
In a nutshell, this study found respiratory microbial co-infections in a whopping 84.6% of severely ill patients, where bacterial and viral co-infections were detected by the aforementioned sequencing approach in 69.2% and 30.8% of the patients, respectively.
Additionally, in 23.1% of the patients, bacterial co-infections with Burkholderia cepacia complex and Staphylococcus epidermidis were confirmed by using classical bacterial culture. One severely ill patient also demonstrated a secondary, time-dependent infection with B. cenocepacia harboring a panoply of virulence genes, which can be linked to disease deterioration and death one month after hospital admission.
Staphylococcus epidermidis bacteri. Image Credit: royaltystockphoto.com / Shutterstock
"Detection and tracking of Burkholderia cepacia complex-associated nosocomial infections are recommended to improve the pre-emptive treatment regimen and reduce fatal outcomes of hospitalized patients infected with SARS-CoV-2", further emphasize study authors.
Moreover, the researchers observed extremely high expression levels of Mycoplasma orale bacterial species in two severely ill patients with prolonged stay in the intensive care unit (i.e., more than 30 days). These results point to the possibility of Mycoplasma-associated co-infection in COVID-19 patients with severe disease, which warrants our increased attention.
"The dominating Proteobacteria transcripts in the gut of antibiotic treatment naïve mild COVID-19 cases in this study, as well as the marked gut microbial changes during respiratory viral infections in prediabetes reported in a recent longitudinal multi-omics study, both support interactions between the host, respiratory viruses and the commensal microbiota", add study authors, corroborating the link between COVID-19 and gastrointestinal symptoms.
Patterns of co-infection informing treatment decisions
This ambitious and intriguing study managed to identify distinct and rather interesting patterns of co-infections. More specifically, not only SARS-CoV-2 was frequently found in correlation with a myriad of respiratory pathogenic microbes in hospitalized COVID-19 patients, but they could also be linked to disease severity.
Moreover, the findings demonstrate the value of using metatranscriptomics for unbiased co-detection of respiratory pathogens, as well as providing timely and useful information for monitoring and managing bacterial co-infections in severely diseased COVID-19 patients.
However, without having pre-treatment specimens, it is challenging to ascertain to what extent differences found in the respiratory microbial patterns (associated with SARS-CoV-2 infections) actually reflect the disease or the antimicrobial treatment – or both.
"Still, our results suggest that the distinct bacterial communities detected in the respiratory and gastrointestinal tract of the severely ill patients might relate to the significant disruption of the normal human microbiota caused by the treatment with antibiotics, allowing colonization by pathogenic antibiotic-resistant bacteria," said study authors in their medRxiv paper.
In any case, the attention of both the global scientific community and public should be directed towards potential future threats from a burgeoning reservoir of antimicrobial-resistant microorganisms and associated genes that will undoubtedly follow the COVID-19 pandemic.
Article source:

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources