A new study by researchers at the University of Oxford and Harvard University and published on the preprint server medRxiv* in July 2020 reports the use of artificial intelligence (AI) to screen patients presenting or admitted to hospital for COVID-19. This could help triage patients in low-testing settings and help reduce infection risk.
The ongoing COVID-19 pandemic is spreading far and wide partly due to the delay in diagnosis, especially when the presenting symptoms are nonspecific. The use of RT-PCR can identify the presence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), but the results may take up to 48 hours. There is, therefore, an urgent need for quick and immediate testing to identify this infection at the point of care. This will expedite required caregiving as well as rapid triage and control of infection.
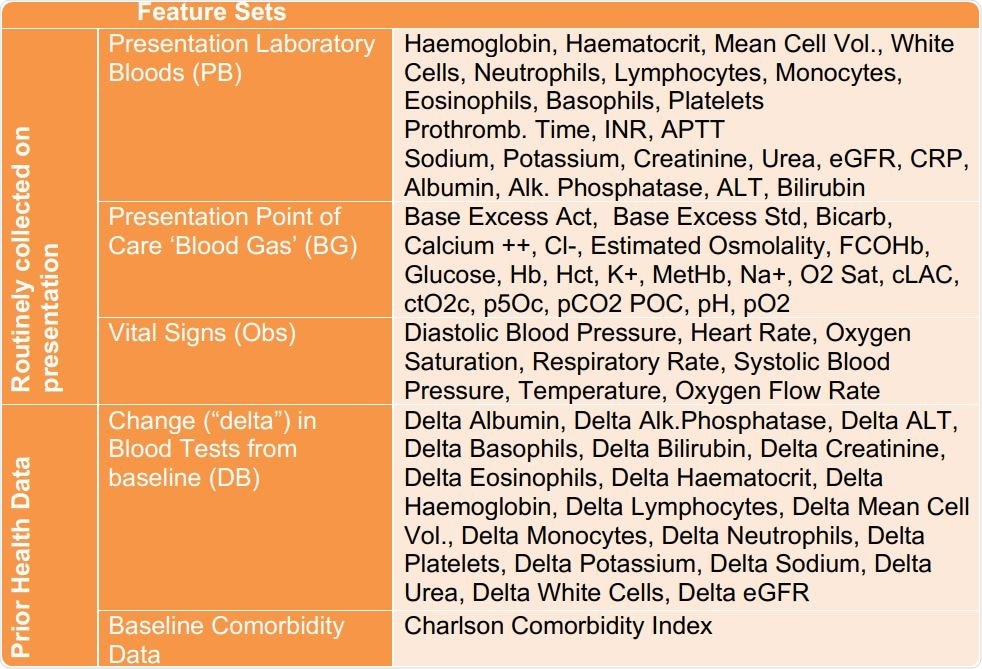
The use of electronic healthcare records (EHR) can be adapted to this purpose, by using machine learning to pick up signs of COVID-19 infection while remaining within the ordinary hospital testing regime. The current study looked at the use of AI trained from clinical data that are routinely collected at hospital presentation, to allow rapid screening for this illness within an hour of being seen by the first healthcare provider.
The study includes more than 170,500 consecutive presentations to either emergency or acute medical departments at a large hospital in the UK. After training the AI machines, they simulated the actual performance of the systems at different points of the epidemic by test sets having a different prevalence of the infection. The researchers examined the predictive values.
They then conducted a validation study on all the patients between April 20, 2020, and May 6, 2020, comparing the predictions to the PCR test results. Almost 115,400 and over 72,300 patients presented and were admitted, respectively.
The researchers developed two models to screen for COVID-19, one in admitted patients and one for all patients presenting at hospital. They found that both models could be trained using routine laboratory blood tests performed at presentation, blood gas results at the point of care, and vital signs. They found that the first and third had the highest predictive value for COVID-19.
The model for detecting COVID-19 among presenting patients had 77% sensitivity but 96% specificity. For those admitted with COVID-19, the sensitivity and specificity was 77% and 95%, respectively.
Over a two week period, these models were prospectively validated against RT-PCR for all patients. They found that the presentation and admission models scored 92% accuracy, with high negative predictive values. In other words, more than 99% of patients for whom the test was negative were truly free of COVID-19.
Implications
The researchers sum up, “Our Emergency Department and Admission models effectively identify patients with COVID-19 amongst all patients presenting and admitted to hospital, using data typically available within the first hour of presentation.”
When the prevalence is low, at 5% or less, these models can achieve a negative predictive value of over 99%, which means they can exclude the disease. When the prevalence is much higher, at above 25%, the models can be adapted to provide a higher positive predictive value of above 83%, which is required. The ability of the models to be adapted to changing conditions as the pandemic continues to spread, and change is an important advantage of this approach.
The high negative predictive values allow their use as a screening test so that patients presenting to emergency rooms can be quickly screened for COVID-19. This will help with giving immediate care, guiding patients through the diagnostic and admission process safely, and even forming a pre-test group for formal RT-PCR when the latter are limited in availability.
The study is important in that it has the largest dataset of any AI-based COVID-19 laboratory study until now, with the large non-COVID-19 patient cohort allowing a broad spectrum of undiagnosed infections and other presentations to be used for training.
Not only does the use of laboratory blood tests and vital signs preclude specialized testing that is not available in the first hour of presentation, but may help identify factors that put individuals at higher risk for COVID-19 or other similar-appearing conditions. Thirdly, this would be ideal for urgent diagnoses in both advanced and poor countries, which have a deficit of testing kits. Finally, they are easily scalable.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Soltan, A. A. S. (2020). Artificial Intelligence Driven Assessment of Routinely Collected Healthcare Data Is an Effective Screening Test For COVID-19 In Patients Presenting to Hospital. medRxiv preprint. doi: https://doi.org/10.1101/2020.07.07.20148361. https://www.medrxiv.org/content/10.1101/2020.07.07.20148361v1
- Peer reviewed and published scientific report.
Soltan, Andrew A S, Samaneh Kouchaki, Tingting Zhu, Dani Kiyasseh, Thomas Taylor, Zaamin B Hussain, Tim Peto, Andrew J Brent, David W Eyre, and David A Clifton. 2020. “Rapid Triage for COVID-19 Using Routine Clinical Data for Patients Attending Hospital: Development and Prospective Validation of an Artificial Intelligence Screening Test.” The Lancet Digital Health, December. https://doi.org/10.1016/s2589-7500(20)30274-0. https://www.thelancet.com/journals/landig/article/PIIS2589-7500(20)30274-0/fulltext.