Tapping into the potential of the multi-omic method to uncover functional connections between various classes of biomolecules, researchers from notable institutions in the United States have discovered sets of co-varying molecules that shed light on coronavirus disease (COVID-19) pathophysiology and provide treatment opportunities. The study can be found on the medRxiv* preprint server.
As of July 2020, the current COVID-19 pandemic (which is caused by the severe acute respiratory syndrome coronavirus 2 or SARS-CoV-2) has resulted in more than 608,000 deaths around the world, primarily due to acute respiratory distress syndrome (ARDS).
As a result of this emerging threat, the scientific response was swift, and many insights into the molecular environment of COVID-19 patients were gained to facilitate the identification of potential therapeutic or vaccine targets.
But regardless of such rapid global reaction to this novel disease, only a handful of studies have explored the broad molecular level reorganization that drives the host COVID-19 viral response.
Luckily, technologies for deep sequence analysis of nucleic acids (often referred to as transcriptomics) are already pervasive. Additionally, high-resolution mass spectrometry can yield similar quantitative data for large-scale lipid, protein, and metabolite measurements.
Considering that extensive profiling across these various planes of biomolecular regulation may provide a holistic view of COVID-19 pathophysiology, a recent paper by the US researchers aimed to leverage these technologies on a large number of patients.
Capturing molecular signatures of COVID-19
In this cohort study, the authors utilized cutting-edge technologies for monitoring thousands of diverse biomolecules from patients with and without COVID-19 in relation to the disease severity and outcomes. And the findings they have obtained are quite intriguing.
The overarching goal of such an approach was to capture the molecular signatures of COVID-19, correlate them with disease severity and clinical metadata, as well as to generate both testable hypotheses and opportunities for therapeutic intervention.
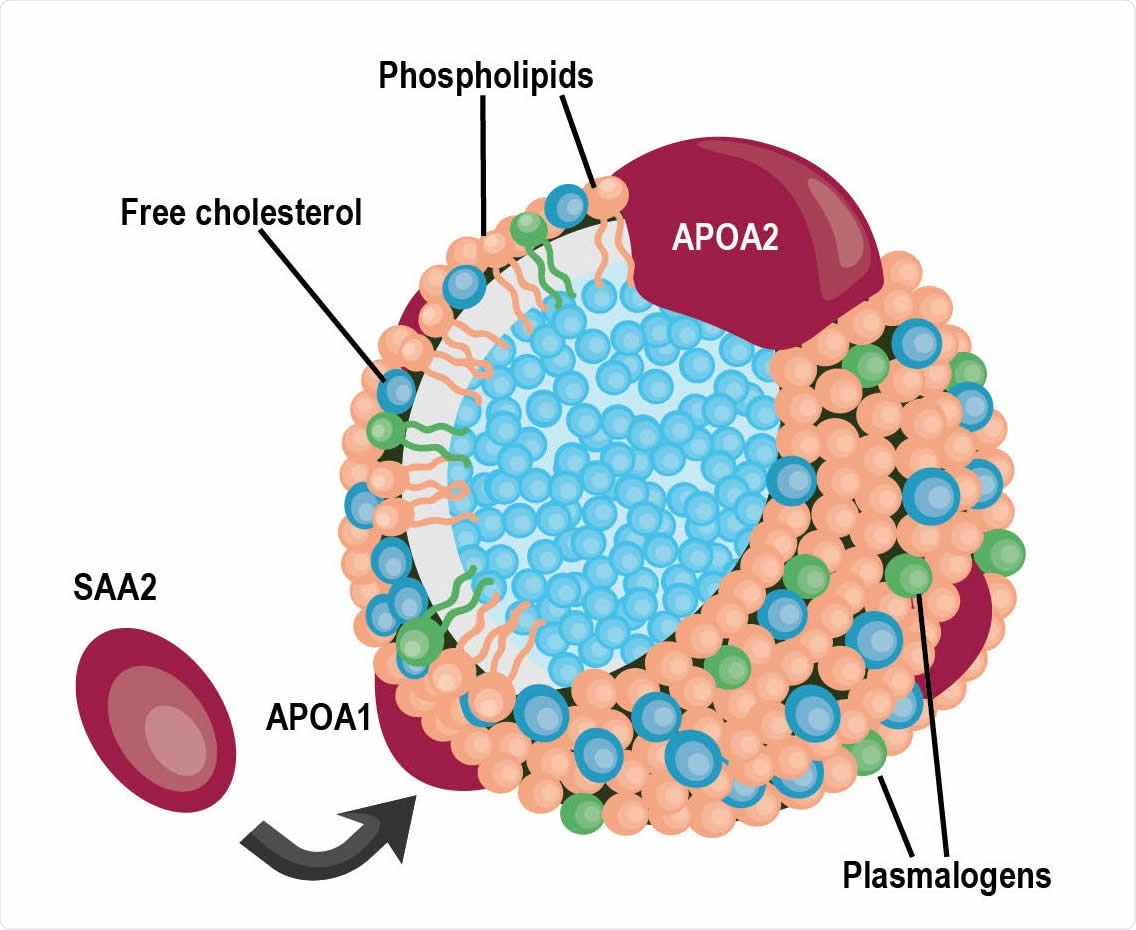
A schematic of a high-density lipoprotein (HDL) particle containing APOA1 and APOA2 proteins surrounded by various lipids, specifically plasmalogens. SAA2, also detected in the cluster in panel b, can replaced APOA1 within the particle.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Consequently, RNA sequencing and high-resolution mass spectrometry have been performed on 128 blood samples from COVID-19 positive and negative patients with varying disease severity.
This resulted in the quantification of over 17,000 transcripts, lipids, proteins, and metabolites, which were associated with clinical outcomes in a curated relational database – uniquely enabling highly specific systems analysis and correlations to molecules and patient prognoses.
Finally, the researchers have decided to make the data broadly available through a free web-based tool (covid-omics.app), enabling interactive exploration of this compendium with the hope that experts around the world will continue to mine these data.
Striking hypercoagulative signature of COVID-19 unveiled
In short, the researchers mapped 219 molecular features with high significance to COVID-19 status and severity, many of them involved in complement and neutrophil activation, as well as dysregulated lipid transport and blood vessel damage.
However, the most striking observations were linked to the dysregulation of blood coagulation and platelet function. As a result, this study provided a unique insight into the COVID-19 hypercoagulation phenotype.
"Our data both confirm the striking hypercoagulative signature of COVID-19 and expand on our current understanding of its pathophysiology", further state study authors. "The data also offer several additional candidate therapeutics," they add.
For example, the authors have observed a substantial reduction in prothrombin abundance and its correlation with disease severity. Furthermore, the levels of cellular fibronectin (which is another coagulation-related protein) were highly increased in COVID-19 patients.
A dysregulation of von Willebrand factor (VWF) (a blood glycoprotein implicated in the cessation of bleeding) and circulating coagulation factors were also observed, which could provide a rationale for more tailored antithrombotic therapies – including the synergistic addition of several drugs that work at different levels.
Precise forecasting with the use of multi-omics based models
"The use of machine learning revealed additional features relevant to COVID-19 severity and underlined the utility of the multi-omics based model for predictions, as this model performed better than the well-established Charlson comorbidity index", study authors summarize their main findings.
Likewise, the addition of the Charlson score (which basically quantifies a person's burden of disease and corresponding one-year mortality risk) as a variable to the proposed model did not result in improved predictive power.
This finding may indicate that the clinical score is highly collinear with the multi-omic variables utilized by this model and that the clinical observation cannot completely capture the features leading to patients' outcomes.
"All large-scale omics studies have limitations but, ideally, still stimulate the generation of numerous testable hypotheses," caution study authors in their medRxiv paper.
This comprehensive work is no exception, and it represents a starting point in our quest to define this devastating disease completely. Future research should, therefore, include a broader and larger patient population, as well as multiple sampling time points.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources