By exploiting the tools of synthetic biology, the researchers from Turkey have successfully developed a de novo riboregulator system for rapidly detecting the presence of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genomic material. Their paper is currently available on medRxiv* preprint server.
The ongoing coronavirus disease (COVID-19) pandemic still poses enormous challenges to societies around the world due to a sheer number of infected individuals. On top of that, SARS-CoV-2 is known to disrupt a wide array of tissues in the human body, sometimes resulting in severe and potentially fatal illnesses.
'Test, trace, isolate' represents a crucial strategy to mitigate the risk of overwhelming the healthcare system until a viable therapeutic option is found. Consequently, widespread testing for COVID-19 is indispensable to halt the viral transmission chain.
Molecular diagnostics of COVID-19 heavily relies on quantitative real-time PCR that identifies the presence of SARS-CoV-2 ribonucleic acid in nasopharynx, trachea, or bronchi; nonetheless, testing capacity often fails to meet increasing demand as a result of the global reagent shortages.
Additionally, real-time PCR tests generally necessitate an adequate testing facility and trained staff. Therefore, in order to compensate for the gap between the pace of infection and test number, many agencies have declared an emergency use of in vitro diagnostic products for SARS-CoV-2.
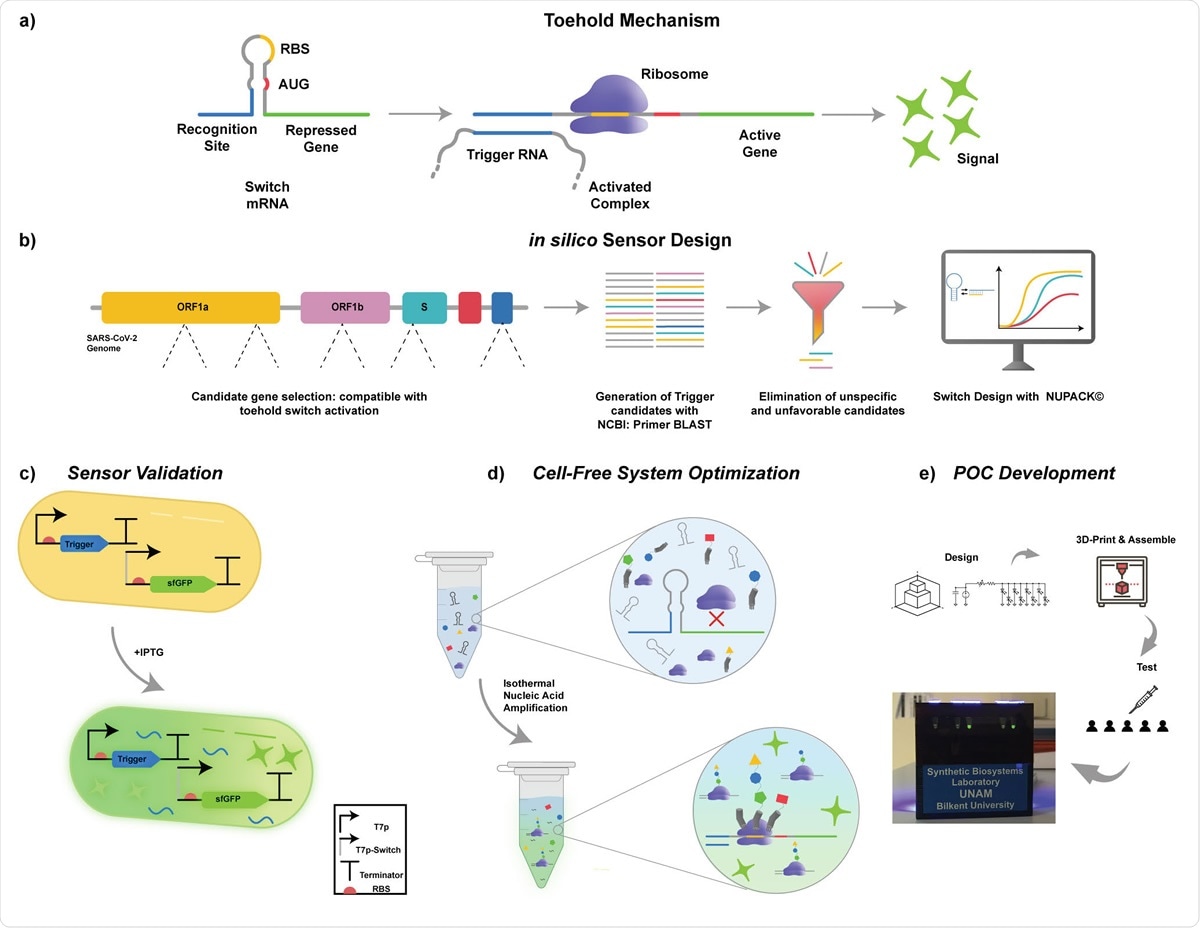
Schematics of the operating principle of toehold biosensors. b) Workflow for cell-free SARS-CoV-2 detection platform. Key target sites for detection of SARS-CoV2 were identified on the viral genome. Toehold triggers are then generated using NCBI Primer BLAST. The trigger candidates that have non-optimal free energy predictions and candidates that are not compatible with species specific amplification were eliminated. Eligible candidates were then used in the design of their respective toehold switches in silico using NUPACK© software. c) Validation of designed toehold switches. Triggers were cloned downstream of T7-LacO promoter. Switches were cloned downstream of T7-LacO promoter and upstream of sfGFP reporter gene. Both constructs were transformed into E. coli BL21 (DE3) cells. Cells that only have the switch plasmid and the cells that have both switch and trigger plasmids were induced with IPTG. d) Cell-free system optimization. e) POC development. Schematics of the development of the portable, low-cost electronic optical reader.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Synthetic biology to the rescue
Synthetic biology was used in the past to broaden the scope of diagnostic tool alternatives, among which versatile and programmable riboregulator toehold switches dominate as a straightforward and inexpensive option. They have already been used for pathogenic viruses such as Ebola and Zika, as well as for appraising antibiotic resistance genes.
In combination with the cell-free transcription/translation (TXTL) technology, toehold switches can be utilized for the detection of specific nucleic acid sequences in point-of-care devices, where the output signal can be a fluorescent protein or enzymatic colorimetric degradation product.
Moreover, the limit of detection of toehold-based sensors can be increased by utilizing simple isothermal amplification reactions, such as recombinase polymerase amplification or nucleic acid sequence-based amplification, prior to TXTL reaction.
This is why researchers from Bilkent University and Ankara University in Ankara, Turkey, decided to design specific toehold switches to detect the presence of the SARS-CoV-2 genome and to establish a proof-of-concept project.
Selecting triggers, constructing switch sequences
More specifically, these researchers selected specific regions in the SARS-CoV-2 genome – ORF1ab, M, ORF678, and S coding sequences – as potential triggers, since they are compatible with the nucleic acid sequence-based amplification reaction.
This was followed by the construction and testing of switch sequences specific to the triggers that control the expression of the superfolding variant of green fluorescent protein. Two out of four trigger sequences (i.e., the regions found in ORF1ab and S proteins) were identified with designed switch sequences.
In short, fluorescence measurements stipulated that triggers selected from a region in S protein and ORF1ab coding sequence may be employed to activate switch repressed translation, and were further characterized in a cell-free TXTL reaction.
Swift recognition of the activated switch
Based on the results of this study, it can be said that the researchers have indeed successfully developed a de novo riboregulator system for detecting the presence of the genomic material of the SARS-CoV-2 virus.
Both trigger/switch pairs displayed similar kinetic properties in the TXTL reaction. The reactions were saturated after 180 minutes when total fluorescence is considered; on the other hand, the 'switch alone' circuit did not yield a meaningful fluorescence signal.
What is the most significant finding for obtaining rapid results in the field, an activated switch could be readily distinguished from inactivated and mock controls even after 40 minutes of the incubation period.
Implementation in a point-of-care device
"Owing to their fast prototyping capability and easy-to-use format, the toehold switch in the TXTL reaction format is an excellent candidate to develop an assay with," study authors emphasize in their medRxiv paper.
"Such assay can be implemented in a point-of-care device for surveillance of SARS-CoV-2 in substantial portions of populations, which do not have access to a centralized facility to perform sophisticated diagnosis assays such as real-time PCR or next-generation sequencing", they add.
In any case, toehold-based biosensors that are implemented in point-of-care devices may indeed facilitate monitoring and COVID-19 transmission control – particularly in rural and less developed regions. Therefore, further characterization assays (which will concentrate on clinical samples) are needed for developing a complete assay as a novel addition to our diagnostic armamentarium.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources