In light of the continuing threat to public health, social functioning, and economic activity posed by the COVID-19 pandemic, scientists are working urgently to bring out a vaccine that is both safe and effective, and capable of mass production. A new paper published on the preprint server bioRxiv* in August 2020 shows that such a vaccine may be produced by using arrays of one severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) protein on a nanoparticle platform.
As a result, the S protein, and particularly the receptor-binding domain (RBD), has been the focus of vaccine development.
Platform Vs. Particle Vaccines
Many novel candidate vaccines currently being developed in response to COVID-19 use platforms to design the antigen, display it, and to deliver the vaccine. The use of such platforms is, however, dependent on the safety, efficacy, and scalability of the vaccine. The first viral vector vaccine has only gained approval less than a year ago.
The current study reports a vaccine produced by the use of self-assembling particulate protein immunogens, which is a technology already in clinical use and demonstrated to be safe and effective in humans. Some previous examples of the use of this technology include the virus-like particle (VLP) vaccines for human papillomavirus (HPV) and hepatitis B virus (HBV). These are recognized to be among the most effective subunit vaccines known.
Computationally Designed Vaccine
Taking this area forward, scientists have found computational design to be an extremely useful platform for the design of protein nanoparticles that display multivalent antigens. Many preclinical studies show that vaccine candidates designed in this manner produce a broader and more potent range of antibody response against many antigens.
The current study describes a self-assembling protein nanoparticle vaccine design based on the structure of the RBD-ACE2 complex. This is termed RBD-16GS-I53-50A, a computationally designed complex of two components, trimeric I53-50A, and pentameric I53-50B. These form an icosahedral complex that is assembled in vitro by mixing I53-50A and I53-50B. The RBD is fused to this complex via glycine-serine linkers.
These constructs were produced within a mammalian cell by recombination. This ensured that the protein was folded correctly and glycosylated. The researchers then tested the antigenicity of the RBD on this platform, and how well multiple epitopes of the RBD in this context were accessible to antibodies. They used two SARS-CoV antibodies, namely, CR3022 and S309, which cross-react with the SARS-CoV-2 RBD.
The researchers found that both these antibodies bound to different epitopes. CR3022 is a conserved hidden epitope that is accessible only when the RBD opens and is not part of the actual receptor-binding motif (RBM), which takes part in the actual ACE2 binding. S309 binds to a glycosylated epitope that is also conserved but is accessible in both the open and the closed conformation. The study shows that RBD-16GS-I53-50 nanoparticles produce the highest binding signals.
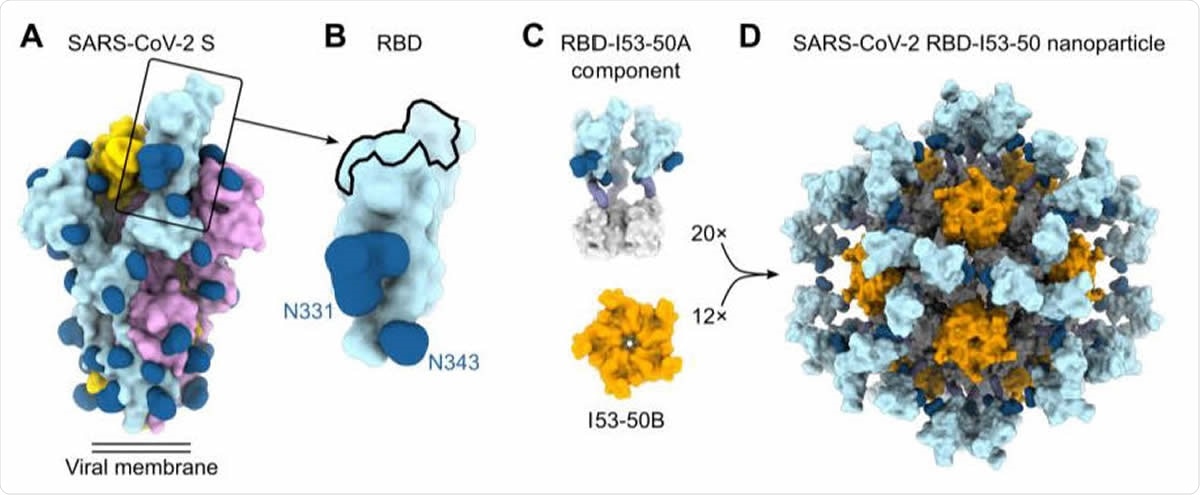
Design, In Vitro Assembly, and Characterization of SARS-CoV-2 RBD Nanoparticle Immunogens (A) Molecular surface representation of the SARS-CoV-2 S-2P trimer in the prefusion conformation (PDB 6VYB). Each protomer is colored distinctly, and N-linked glycans are rendered dark blue (the glycan at position N343 was modeled based on PDB 6WPS and the receptor-binding motif (RBM) was modeled from PDB 6M0J). The single open RBD is boxed. (B) Molecular surface representation of the SARS-CoV-2 S RBD, including the N-linked glycans at positions 331 and 343. The ACE2 receptor-binding site or RBM is indicated with a black outline. (C) Structural models of the trimeric RBD-I53-50A (RBD in light blue and I53-50A in light gray) and pentameric I53-50B (orange) components. Upon mixing in vitro, 20 trimeric and 12 pentameric components assemble to form nanoparticle immunogens with icosahedral symmetry. Each nanoparticle displays 60 copies of the RBD. (D) Structural model of the RBD-12GS-I53-50 nanoparticle immunogen. Although a single orientation of the displayed RBD antigen and 12-residue linker are shown for simplicity, these regions are expected to be flexible relative to the I53-50 nanoparticle scaffold.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Stability and Immunogenicity of the Vaccine
The subunit vaccines are found to be very stable, both physically and in relation to their antigenicity, and are more stable than the SARS-CoV-2 prefusion-stabilized S (S-2P) trimer. This makes them suitable for mass production, safe storage, and distribution without loss of potency. Moreover, the yield of these particles is much higher than for the S-2P trimer.
The researchers say, “The high yield and stability of the protein components and assembled nanoparticles, especially compared to the SARS-CoV-2 prefusion-stabilized S trimer, suggest that manufacture of the nanoparticle vaccines will be highly scalable.”
Though the RBD monomer does not stimulate a potent immune response by itself, the current study also showed that in multivalent form it can be part of a very immunogenic vaccine. This is in agreement with earlier research, which shows that dimerization of the RBD makes it much more immunogenic.
The current vaccine is therefore designed to display 60 copies of the RBD, producing a highly potent array of immunogenic protein, on a self-assembling scaffold. In an animal model, the vaccine was found to induce a powerful neutralizing antibody response, more than the S-2P trimer stabilized in a prefusion conformation. When two doses were given in human subjects, the neutralizing antibody response was higher than that produced by infection with SARS-CoV-2.
Interestingly, either the 0.9 and 5 μg doses of the vaccine produced high and comparable titers of neutralizing antibodies. This means that lower doses are adequate for vaccine efficacy, which is an important aspect when it comes to universal immunization requiring mass-scale vaccine production. The researchers comment, “Both enhanced potency and dose-sparing could be critical for addressing the need to manufacture an unprecedented number of doses of vaccine to respond to the SARS-CoV-2 pandemic.”
Targeting Multiple Epitopes
The study also demonstrates that the vaccine produced a range of antibodies against multiple distinct epitopes, which could explain why it generates such potent neutralizing activity. Moreover, such a polyclonal response minimizes the risk of emergence or selection of escape mutations. Such antibodies are, therefore, a sign of long-term vaccine efficacy.
Secondly, these antibodies are of higher quality than those that are generated by immunization with the S-2P trimer, or by natural infection, because they target many and different epitopes on the RBD that are bound by most neutralizing antibodies. The high proportion of neutralizing to binding antibodies also means that the vaccine is unlikely to produce antibody-dependent enhancement of disease.
The researchers sum up, “These results highlight the utility of robust antigen display platforms for inducing potent neutralizing antibody responses and have launched cGMP manufacturing efforts to advance the lead RBD nanoparticle vaccine into the Continued development of such technology platforms could lead to vaccines that prevent the next pandemic rather than respond to it.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources