The virus behind the current COVID-19 pandemic, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is known to spread more efficiently than the earlier pathogenic coronaviruses, SARS-CoV, and MERS-CoV. However, the case fatality rate so far has been much lower, at 2% to 5%, compared to 10% in SARS and ~ 40% in MERS. Scientists think the virus is inhibited by interferons (IFNs), even more than the earlier viruses. In fact, IFNs are currently being used to reduce the severity of COVID-19.
.jpg)
This scanning electron microscope image shows SARS-CoV-2 (round magenta objects) emerging from the surface of cells cultured in the lab. SARS-CoV-2, also known as 2019-nCoV, is the virus that causes COVID-19. The virus shown was isolated from a patient in the U.S. Image captured and colorized at NIAID's Rocky Mountain Laboratories (RML) in Hamilton, Montana. Credit: NIAID

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
What are IFITMs?
Interferon-induced transmembrane proteins (IFITMs 1, 2, and 3) are proteins that are considered to be inhibitory of a variety of viruses, including the SARS-CoVs. Most of the evidence for this has come from studies that used cells that overexpress these proteins and are infected by pseudoviruses.
The investigators looked at innate immune effectors directed against SARS-CoV-2 entry into the target cells. Viral entry involves spike-mediated recognition of the host receptor, angiotensin-converting enzyme (ACE) 2, triggering an irreversible conformational change of the spike protein to its fusion form by proteolytic cleavage into S1 and S2 subunits. The cleaved protein fuses to the plasma membrane and gains entry to the cell.
IFITMs are a family of IFN stimulated genes (ISGs) is known to prevent this fusion, in the case of influenza A viruses, rhabdoviruses, and HIV.
IFITM Overexpression Inhibits Pseudovirus S Binding
Previous work has shown that when these proteins are expressed at excessively high levels, pseudoparticles expressing the spike protein of SARS and MERS are unable to enter the host cell. The mechanism of inhibition might reduce the rigidity and curvature of the plasma membrane such that fusion cannot happen. While IFITM1 is only the plasma membrane, IFITM2 and 3 are localized on lysosomal membranes within the cell. Many scientists think that such viruses cannot replicate in cells where these proteins are expressed.
However, some studies have shown that IFITMs can actually increase the intensity of infection with some human coronaviruses. At the same time, mutant IFITMs could enhance infection with many viruses from this family, including SARS.
The current study shows that the overexpression of IFITMs specifically reduces the entry of SARS-CoV-2 spike-mediated pseudoparticles, by two orders of magnitude for IFITM2 and IFITM3 in particular, and less potently by IFITM1. Infectivity of these pseudoviruses was not reduced, however, and in fact, it was slightly increased in one case, since it may increase the rate at which the spike protein is built into the pseudovirus.
The initial tests showed that both SARS-CoV and SARS-CoV-2 spike proteins expressed in pseudoviruses are inhibited efficiently by IFITMs, the first even more than the second. The mechanism of such inhibition appears to be via ubiquitination and palmitoylation.
In all cases, they found that IFITMs reduce cell-to-cell fusion mediated by the spike-ACE2 binding. The depletion of these proteins led to a 3- to 7-fold increase in spike-mediated infection by all pseudoviral particles. Further testing in a cell line lacking IFITM expression showed that the number of S-ACE2 binding foci leaped upward by four- to ten-fold. These findings strongly imply that IFITM proteins are efficient inhibitors of SARS-CoV-2 S-mediated viral entry.
The question is whether this susceptibility to IFITM-induced inhibition of S-ACE2 binding is shared by SARS-CoV-2 even after its leap across the species barrier to infect human hosts.
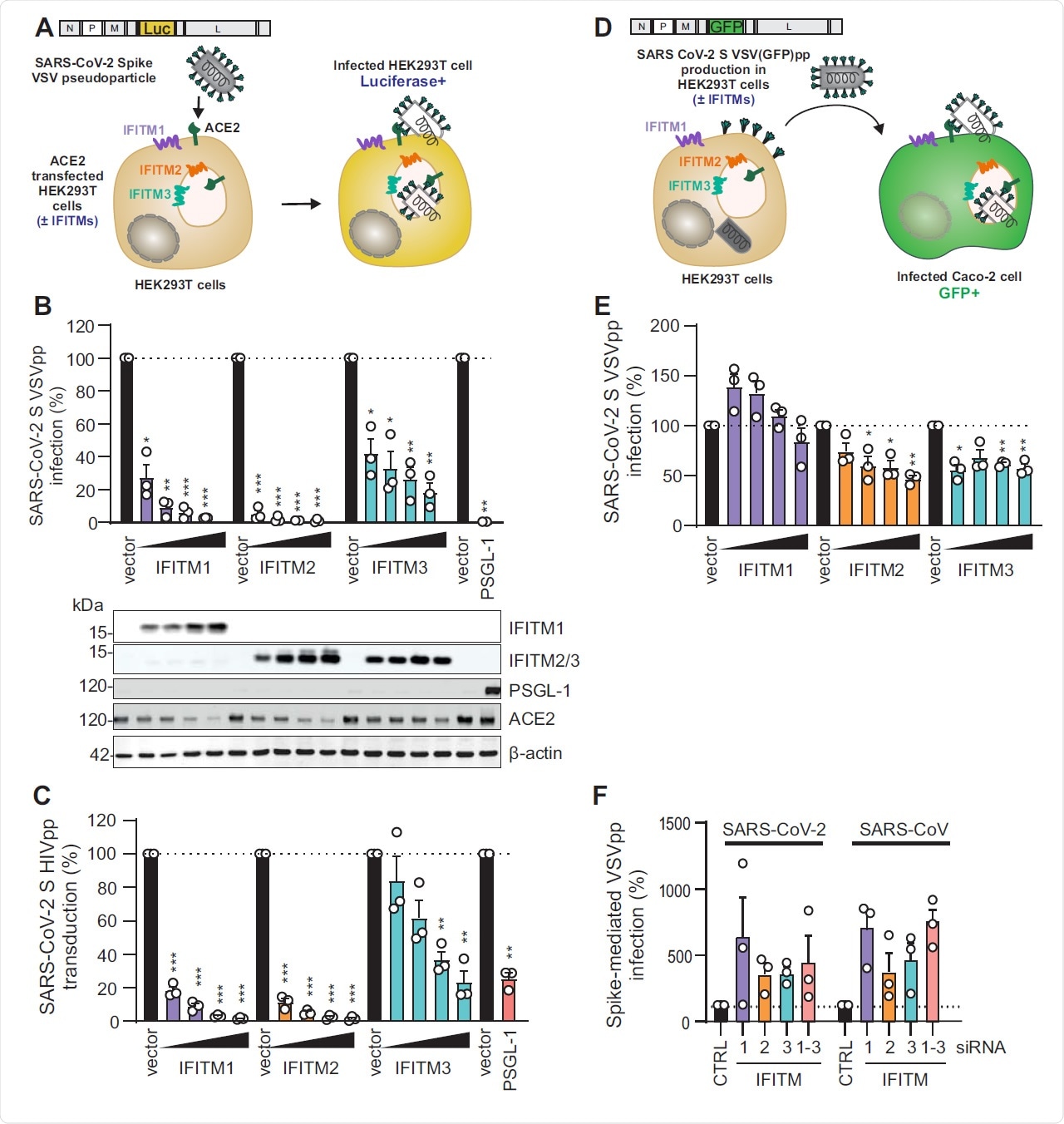
Figure S1 (related to Figure 1). VSV-G-mediated infection by VSVpp is not significantly inhibited by IFITM proteins. (A) Quantification of VSV(luc)ΔG*VSV-G entry by luciferase activity in HEK293T cells transiently expressing indicated proteins and infected 24h post transfection with VSV(luc) ΔG*VSV-G (MOI 0.025) for 16 h. Bars represent means of n=3±SEM. Lower panel: Immunoblot of the corresponding whole cell lysates (WCLs) stained with anti-IFITM1, anti- IFITM2, anti-IFITM3, anti-PSGL-1, anti-ACE2 and anti-actin. (B) Immunoblot analysis of whole cell or supernatant lysates of HEK293T cells transiently transfected with SARS-CoV-2 S VSVpp and increasing doses of IFITM1 expression construct. Blots were stained with anti- Spike, anti-VSV-M, anti-IFITM1 and anti-actin. (C) Quantification of VSV(luc) ΔG*VSV-G particles by luciferase activity in Caco-2 cells infected with the supernatant from HEK293T cells transiently transfected with VSV-G and empty control or IFITM expression vectors. Bars represent means of n= 3±SEM. (D) Exemplary immunoblots of whole cell lysates of Calu-3 cells transiently transfected with control siRNA (si.NT) or siRNAs targeting IFITM1, 2 or 3 as indicated. Percentages indicate signal intensity of the three IFITM proteins relative to those observed in the presence of the control siRNA (100%).
IFITM Reduces Viral Loads
The scientists found that in response to interferons, IFITMs are produced in the lung, which is the primary site of infection by SARS-CoV-2. It is also induced in other infected tissues. IFNs have both positive and harmful effects on the tissues, and may even worsen the disease and trigger severe COVID-19. They also activate a cascade of pro-inflammatory cytokines and chemokines, with as yet unknown effects on the disease.
The experiment was repeated using authentic SARS-CoV-2 on a cell line expressing the ACE2 receptor alone or along with IFITMs. They found that when IFITM1 or 2 was overexpressed, viral RNA loads decreased dramatically by over 30-fold and 136-fold. However, IFITM3 produced a fivefold inhibition.
Endogenous IFITM Enhances SARS-CoV-2 Infection on Calu-3 Cells
IFITM2 AND 3 are expressed constitutively at low levels on primary human lung bronchial epithelium, as well as on Calu-3 cells which express both ACE2 and all the IFITMs. The expression of all IFITMs was enhanced by treatment with IFNs, but to different levels depending on the cell type. IFN-β and IFN-γ were most efficient at increasing IFITM1 and IFITM2 in bronchial epithelial cells and IFITM1 in Calu-3 cells, respectively, while IFN-β was active in intestinal organoids.
The investigators tested the effect of IFITMs on the Calu-3 cell line. They found that when the wildtype virus was used to infect a human lung cell line, Calu-3, endogenous IFITM expression enhanced the infection, both with and without interferon. The most important IFITM for this process was IFITM2, which increased viral entry and production by several orders of magnitude. However, IFITMS failed to enhance SARS-CoV-2 infection in HEK cells at different levels of expression.
When these endogenous proteins were silenced by siRNA knock-down, authentic SARS-CoV-2 replication took a steep nosedive, by 3-4-fold with IFITM1 and IFITM3, and by 22-fold when IFITM2 was absent. At 24 hours, IFITM2 reduced the viral load by over two orders of magnitude.
Thus, the endogenous expression of IFITMs does not restrict but rather increases the entry and replication of this virus in human lung cells. Treatment with IFN-β caused a 22-fold reduction in viral RNA levels at 48 hours from infection while silencing the IFITMs further reduced them by 4- to 68-fold.
The infectious titer was examined by cytopathic effect (CPE) assay on Vero cells. This confirmed a radical decline in infectious potential upon silencing all the IFITMs. If only IFITM1 or IFITM3 was depleted, the CPE was modest and occurred only at low dilutions. With the depletion of IFITM2, CPE was abolished, proving that the lack of this protein reduced the yield of infectious viral particles by over four orders of magnitude.
Implications and Future Directions
They concluded that IFNs enhanced the baseline expression of IFITM proteins in lung cells and other tissues infected by SARS-CoV-2, enabling viral spread in vivo. Calu-3 cells are, therefore, a good model for studying this infection.
In other words, the researchers say, “IFITM expression is critical for efficient SARS-CoV-2 replication in Calu-3 cells. [The virus uses this protein] to achieve efficient replication and spread under physiological conditions.’ This may also contribute to severe COVID-19 since the antiviral factors allow the virus to infect the cells of the lower respiratory tract efficiently.
This study also shows how essential it is to confirm observations of antiviral effects under in vivo conditions. Previous investigators may have missed the enhancing effect because it is specific for endogenously affected IFITMs in human lung cells infected with the wildtype virus. Antibodies to these proteins may be an interesting and novel therapeutic avenue for this infection.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources