As the COVID-19 pandemic in the US continues to spread aggressively, testing remains somewhat dysfunctional and uncoordinated. This has made it challenging to test for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and to obtain results in time for action to be taken to contain the spread of the virus. Now, a new study by researchers at Yale School of Medicine and Yale School of Public Health and published on the preprint server medRxiv* in September 2020 shows that pooled saliva samples can save resources and increase testing capacity, making a significant difference in the management of the pandemic.
The most significant constraint in stopping the virus has been the limited availability of tests and testing kits, reagents, and laboratories. However, testing is crucial to not only diagnose but also to screen for the disease and to monitor and predict outbreaks. To increase the testing capacity in all places where gatherings of people are necessary for the return to near-normal life, such as schools and workplaces, pooled samples have become the way to go.
Also called batched testing, the pooling of test samples allows multiple people to be tested together to detect the presence of the virus in one or more of the samples. If the result is negative, all the individuals are reported to be negative, while if positive, each sample must then be individually re-tested.
So far, pooled samples have used reverse transcriptase-polymerase chain reaction (RT-PCR) to detect viral RNA, after first extracting it from individual samples. However, pooled testing can be initiated after mixing the samples and extracting the pooled RNA just as well. Earlier studies have shown that these approaches save resources, but laboratory data is still necessary to back up future modifications.
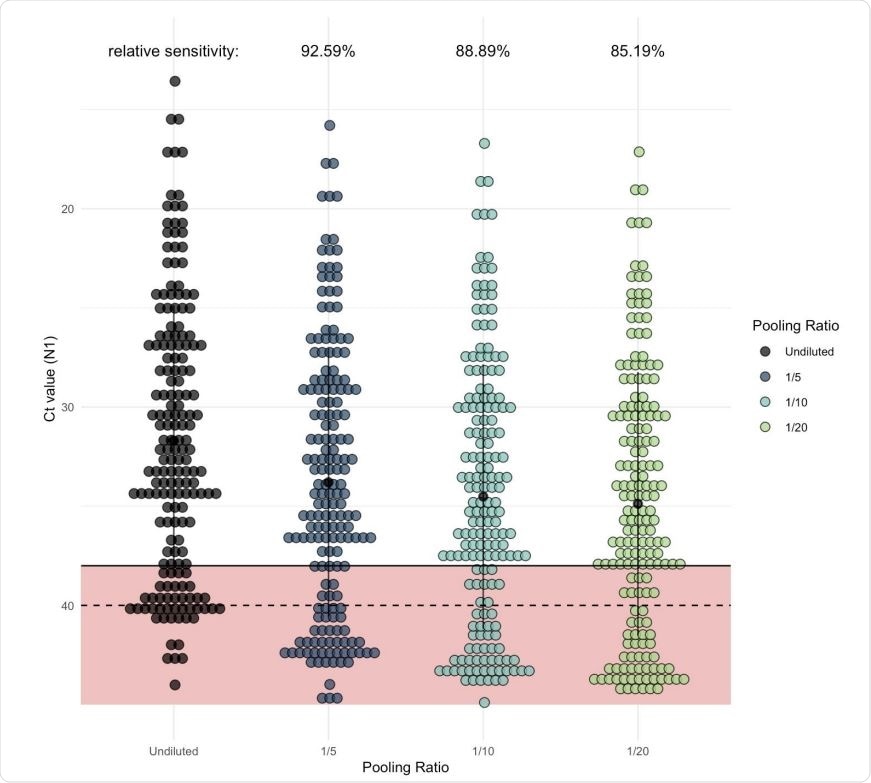
As pool size increases, more samples would be classified as negative in comparison to samples tested individually (unpooled). Each dot represents one of the Yale IMPACT saliva samples which generated signal when tested by RT-qPCR for SARS-CoV-2 N1. Of these, 135 fell below the cycle threshold (Ct) of 38 and were classified as positive for virus. The regression coefficient (representing expected Ct increase) for each of the pool sizes was added to the Ct value generated from the undiluted sample (shown in black) to determine the relative level of sensitivity for each pool size.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Saliva for Viral RNA Detection
Recently, saliva has been shown by the current team of researchers to be an efficient option for viral RNA detection. Not only is it easy to self-collect, but it does not require either stabilizing buffers or cold chain maintenance during transport.
Different Pool Sizes
The current study focuses on pooling 5, 10, and 20 saliva samples for a high-throughput testing workflow. The samples came from COVID-19 inpatients and healthcare professionals. The samples containing the virus were combined for all cycle threshold values below 38 Ct with negative saliva. Subsequently, they performed RNA extraction and detection.
All samples included in the pool had already been tested individually. The comparison between the pool testing results and the latter shows that with increasing pool size, test sensitivity declined by ~2, 3, and 3.6 Ct with a pool of 5, 10, and 15, respectively. This effect does not vary with the Ct value for the undiluted sample, and therefore will not cause a decline in sensitivity at different Ct values. In other words, it remains equally sensitive at various viral loads.
These results agree with previous pooled swab testing research findings. However, the current study also used increased extraction volume, from 300 μL to 400 μL, but without changing the elution volume, to enhance sensitivity to a level comparable to that for undiluted samples.
Again, the effect of pooling the samples after extraction of RNA, and of using pooled RNA templates from undiluted saliva samples, in sets of 5 and 10, showed the decrease in sensitivity to be similar to that obtained with pre-extraction pooling. However, with the latter, the individual sample variance was smaller.
Analysis of the Ct values for all positive samples from their previous study showed that with a pool of 5, 10, and 20 samples, about 93%, 90% and 85% of samples would be correctly detected, respectively, in comparison to the individual detection of the samples. In other words, the sensitivity reduces by 12-15% with pooled testing relative to individual testing.
The Cost-Effectiveness of Pooled Saliva Testing
A broad-based testing technique must also be inexpensive in order to achieve its objective of identifying infectious people to isolate them from the population. The cost-effectiveness of this strategy was, therefore, compared to individual sample testing. The researchers modeled the number of tests required for a population of 10,000 at varying levels of prevalence, using pools of 5, 10, or 20.
This showed that the number of positive pools would increase with rising prevalence, which means more individual tests will be required to confirm the presence of the infection in individuals.
Reduced Number of Tests
When the prevalence is 3% or more, therefore, smaller pools of 5 will result in the optimal number of tests overall. But with lower prevalence, larger pool sizes of 10 or 20 will reduce the number of tests required, saving on both cost and test supplies. For instance, with a prevalence of 0.8%, a pool size of 20 would be ideal to ensure that ongoing population surveillance and early identification of new outbreaks are feasible.
In general, once the tested population has a prevalence of 30% or less, pooled samples would reduce the number of tests needed. Beyond this prevalence, pooled sizes were not tested. However, with reduced prevalence, the cost savings associated with pooled testing increases.
To illustrate this, the researchers found that with a prevalence of 0.5%, just over 1,300 tests would be enough to cover a population of 10,000 people. This would mean saving over $260,000 by pooled testing vs. individual testing, given that tests typically cost $30 each, while still identifying 43-50 infections.
Increased Testing Frequency
An even greater advantage is that increased testing frequency is allowed, which means more infectious individuals are identified and isolated, preventing viral spread. Thus, even in areas of low prevalence, pooled testing still has significant economic benefits, promoting continued surveillance.
The researchers comment, “Our model demonstrates that as local outbreaks fluctuate, varying pool sizes in response will have resource-savings benefits. Taken together, pooled testing of the non-invasive and cost-effective saliva sample type facilitates extended duration and breadth of screening test strategies.”
Screening for surveillance, not Diagnosis for Treatment
The pooled testing strategy approved by the US Food and Drug Administration (FDA) will be of greatest value in a population with a high prevalence of infection. However, since the sensitivity drops by ~12-15% with pools of 10 or 20 samples, FDA authorization may occur only if this is recognized to be a screening strategy rather than a diagnostic tool, and if the capability to offer more frequent and repeated testing is also considered.
Again, they say, “The probability of a false negative should not be considered per test, but rather for a given testing regime over a specified period of time.” They also consider the real number of tests required to be probably still lower, as there are likely to be more than one positive result in a single pool. Test protocols can also be tweaked to yield the most significant number of true positives by reducing the effect of sample dilution. For instance, the sample volume could be increased to the maximum, or the RNA elution volume reduced, while an order of magnitude increases the Ct positivity threshold. The RNA extraction step may also be omitted until after pooling.
The study concludes, “Together with the ease of saliva collection, this strategy should be considered as an effective testing strategy to expand the breadth of testing and continued surveillance during the ongoing COVID-19 pandemic.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources