The SARS-CoV-2 is a single-strand betacoronavirus, with a large genome encoding both structural and non-structural proteins, the latter being concerned with viral replication and other viral functions. The structural proteins include the spike (S) protein, the nucleocapsid (N) protein, the envelope (E) protein, and the membrane (M) protein.
The complete virus requires all four of these proteins to be present. The S protein mediates viral attachment to the host angiotensin-converting enzyme 2 (ACE2) receptor, and then the fusion of the viral and host cell membranes, with viral cleavage, that allows the virus to gain entry into the cell and initiate active infection.
The N protein is among the most abundant, and its binding to the RNA genome triggers replication and assembly of the viral proteins, as well as initiating the host cellular response during viral infection.
Mutagenesis in SARS-CoV-2
All organisms undergo mutation, which is often or mostly harmful to the organism but also drives natural changes in the adaptation of the organism. This also applies to the SARS-CoV-2 genome. The researchers in the current study used the SARS-CoV-2 Mutation Tracker to examine the mutational history of the virus.
They found that almost 15,000 mutations have occurred so far, with over a thousand being on the S gene. This impacts the infectivity of the virus and must be considered in any evaluation of viral spread, because different geographic and demographic characteristics, as well as exposure to a wide range of factors that adversely affect the viral genome, drive these mutations.
Research shows that the virus is mutating at a slower rate than other common viruses such as the common cold and influenza viruses. This is attributed to the efficient proofreading apparatus in the coronavirus genome, especially those in the order Nidovirales, which involves the function of a non-structural protein (nsp) 14 along with nsp 12 (RNA-dependent RNA polymerase, RdRp).
Mutations in the SARS-CoV-2 arise from three routes: random replication errors, due to genetic drift and exposure to spontaneous genotoxins; defects in the replication, proofreading, and repair mechanisms allowing mistakes to persist; and immune responses by the host cells that destroy the gene integrity of the virus.
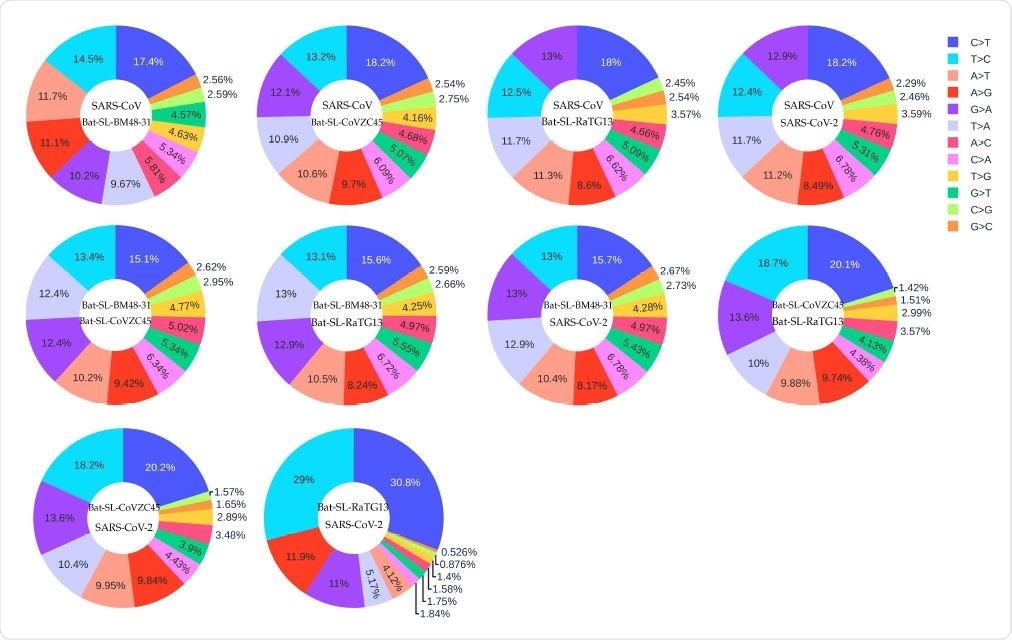
The distribution of 12 SNP types among SARS-CoV, Bat-SL-BM48-31, Bat-SL-CoVZC45, Bat-SL-RaTG13, and SARS-CoV-2. Here, the text on the top represents the reference genome and the text at the bottom represents the mutant sequence.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Genotyping to Track Mutational Variants
One method to understand this is by genotyping, which allows researchers to trace the course of the mutation over a population, time, and space, as well as to examine how the viral proteins actually work. As of now, most genomic studies on this virus have targeted different variants of the genome and how they affect viral spread as well as diagnostic methods, vaccines, antivirals, and antibodies.
Hypermutations in SARS-CoV-2
Earlier studies have shown that the Wuhan strains of the virus show hypermutations C>T occurring as a result of deamination during RNA editing as a result of APOBEC (apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like) activity. TAA, TAG, and TGA are the three stop codons in the standard genetic code. Therefore C>T mutations will probably result in premature termination of viral protein translation, causing the viral proteins not to be produced or to be non-functional or poorly functional. This, in turn, impairs viral survival.
RNA Editors in Human Cells
The two RNA editing mechanisms known in human cells are deaminases, namely, the RNA-APOBEC and RNA-ADAR (adenosine deaminases acting on RNA) editing mechanisms. The first of these replaces cytosines to uracil (C>U) on single strands of nucleic acid. The latter changes adenines into inosines (A-to-I) abundantly in both viral and host sequences, and result in A>G mutation.
The human genome also encodes activation-induced cytidine deaminases (AIDs) as well as several other APOBEC homologs for cytidine deamination, which are active in innate immunity and RNA editing. They cause mutations in specific sequences of the genome of both hosts and pathogens.
Why Hypermutations Occur
These mechanisms cause spontaneous C>T and A>G mutations, but the very high proportions of these mutations indicate that they are part of the host reaction to the virus. A random mutation rate will result in a ratio of ~8% per mutation, but the C>T mutation accounts for ~24% of mutations – hypermutation. The A>G mutation is also present at a frequency of ~15%. This indicates that additional mutations are at work
This indicates that the host immune system is fighting the virus via its RNA editing mechanisms. However, the virus counter-attacks, using its own proofreading and repair mechanisms, accounting for the high reversed mutations T>C and G>A.
Mutations can be classified into the four transition types mentioned above, and eight transversion types. However, the latter is relatively low frequency except for G>T, probably due to the ease with which single ring nucleotides can be substituted for each other and the increased risk of damaging mutations.
Age Associated Hypermutations
C>T mutations are far more common with age; over 42% of them being found in patients over 90 years of age. This may mean that their immune responses are more aggressive, or due to unregulated responses as seen in the cytokine storm often seen in severe COVID-19. This phenomenon often causes inflammation and apoptosis to increase in an exponential fashion, causing organ damage. This may account, therefore, for the higher mortality due to COVID-19 in the elderly.
The C>T mutation rate is also comparable in children below 5 years of age. Notably, these two age groups also have the second-highest ratio of T>C reversed mutations. This indicates that viral counter-attack will also be higher in these individuals.
The researchers suggest that in children younger than 5 years of age, the immune system is immature and weaker, which could put them at risk of more severe COVID-19, but more work is needed to reveal the long-term consequences of this infection on their health.
Females also have a slightly higher ratio of C>T mutations, indicating that they have a more robust immune response, except in those aged 6-19 years and over 90 years.
Geographic Analysis
The researchers also found that the highest C>T mutation ratios are in the UK isolates, compared to Australia, India, and the USA. However, older patients in the UK and Australia lack the otherwise consistent global increase in the C>T ratio in this age group.
Isolates from Europe, Asia, and North America show a lower C>T mutation ratio, below 35%, and a higher T>C ratio, over 10%, but the reverse is found in Oceania, Africa, and South America. In fact, the latter have C>T 10% or more higher than the former, possibly indicating a more robust immune response with more active APOBEC editing in the latter.
Oceanic and African isolates also have unusually low A>G mutation ratios less than 9%, unlike other regions. This may indicate that Asian and European populations have different immune responses to the virus, in terms of genetic and molecular mechanisms.
Preferential SNPs
The researchers also looked at almost 14,000 SNPs derived from over 33,000 isolates to analyze the mutation frequencies of C>T, T>C, A>G, and the reverse mutation, in 2-mer and 3-mer sequence contexts. They found specific mutation patterns depending on the position of the SNP in the 2-mers and 3-mers, and were able to decipher the preferred genetic environment of the virus.
Comparing Mutations Between Coronaviruses
The researchers assumed that the five coronaviruses in this study have the same ancestral strain. They found that they all had high rates for the four transition mutations, showing that the APOBEC (C>T) and ADAR (A>G) gene editors are driving the RNA changes among and within these strains.
They speculate that based on the C>T to T>C ratio, these viruses may be ranked in order of emergence from the common ancestral strain.
Implications
The study indicates the vital role played by the APOBEC proteins in innate and adaptive antiviral responses early in the course of the infection. A higher frequency of the hypermutation C>T demonstrates strong host immunity, leading to efficient viral clearance. However, this may turn hyperactive and lead to host tissue or organ damage, and death, in COVID-19, by triggering a cytokine storm.
The researchers conclude, “We hypothesize that virus genomes evolve through host innate immune response-imposed gene editing, i.e., C>T, and virus protective mechanism-installed defective revisionary mutations, T>C. As a result, both C>T and T>C mutation ratios are usually high.” However, the higher frequency of the former indicates that it is the driving force to which the latter is the response – a “master and slave relationship.”
This finding can be used to detect the direction of variation, in that, a C>T to T>C ratio higher than 1 indicates a forward trajectory.
Finally, this study throws light on the recovery of some infected individuals without detectable humoral immunity, which could be due to strong APOBEC3 activity that cleared the virus without the production of neutralizing antibodies.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources