Although 3p21.31 is known to be a major genetic locus involved in severe COVID-19, until now, the underlying pathological mechanisms and target genes involved have remained unknown.
Now, Kaixiong Ye and colleagues have shown that this COVID-19 risk variant is associated with altered blood cell traits involving monocytes, eosinophils, and neutrophils, and the expression levels of their candidate target genes CCR1 (C-C chemokine receptor type 1), CCR2, and CCR3.
The researchers say the multiple blood traits identified in this study, as well as their candidate acting genes, represent intriguing and testable hypotheses that should be followed-up in future studies for the role they play in COVID-19.
A pre-print version of the paper is available on the medRxiv* server, while the article undergoes peer review.
Genetic variation plays a role in the different responses people have to infection
The clinical manifestations of COVID-19 vary significantly between individuals, ranging from asymptomatic illness to flu-like symptoms, loss of smell, pneumonia, and respiratory failure.
Aside from underlying health conditions and demographic factors such as older age and male gender, genetic variation also plays a role in the different responses people have to infection.
The first genome-wide association study (GWAS) to assess COVID-19 among patients who developed respiratory failure identified genetic associations at the locus 3p21.31. This finding was replicated by the COVID-19 Host Genetics Initiative.
“However, the underlying causal variant, the target gene, and the pathophysiological process are unknown,” said Ye and colleagues.
The PheWAS approach
Phenome-wide association study (PheWAS) is an approach that can be used to evaluate the associations between a disease-related genetic variant and a wide range of phenotypes. This method can identify intermediate traits or biomarkers involved in the causal pathway, from the genetic variant to the disease of interest.
Now, Ye and colleagues have performed a PheWAS to investigate the intermediate traits underlying the link between locus 3p21.31 and severe COVID-19.
The team leveraged deep phenotyping information available for 310,999 European individuals in the UK Biobank and performed a PheWAS for a COVID-19 risk allele with 923 disease phenotypes, biomarkers, and blood cell traits. Candidate associations were then replicated in another dataset.
Next, for candidate genes potentially regulated by the COVID-19 risk variant, associations between their expression levels and the polygenic scores (PGS) of 1,263 traits were evaluated in a meta-analysis of 31,684 blood samples.
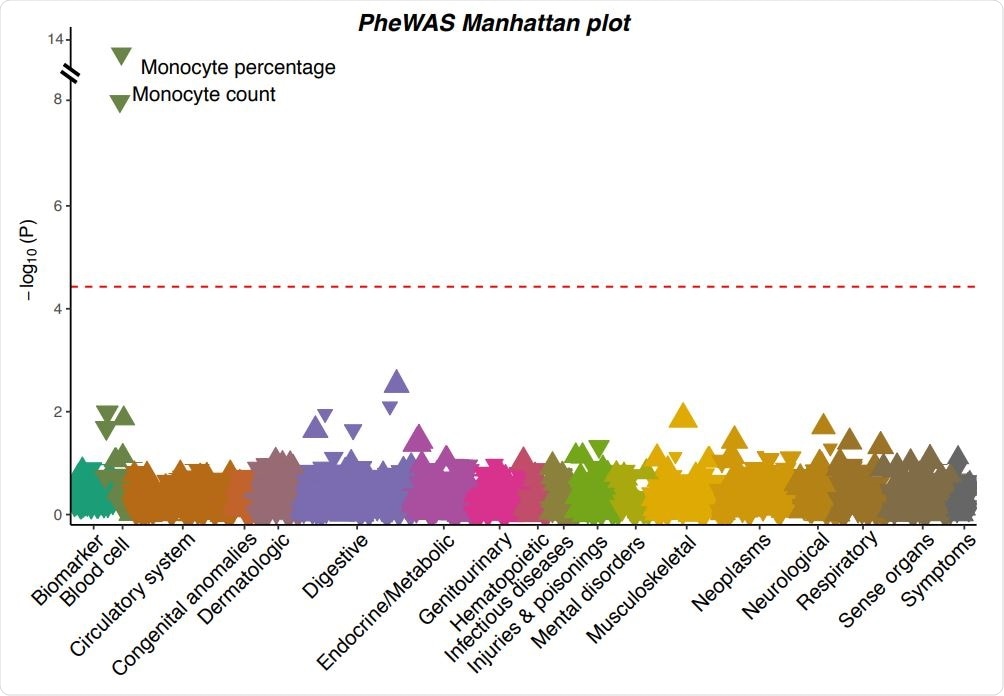
A Manhattan plot showing the associations between the severe COVID-19 risk variant and 923 phenotypes in UK Biobank. Each triangle represents one phenotype. Triangles pointing up indicate increasing effects of the COVID-19 risk allele on the phenotypes, while those pointing down indicate decreasing effects. The size of the triangle is proportional to the effect size. The significance threshold with Bonferroni correction (p < 0.05 / 923 = 5.42×10-5 ) is represented by the red dashed line.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The variant is associated with altered blood traits
The PheWAS in the UK Biobank for the severe COVID-19 risk variant, along with replications identified in an additional dataset, revealed that the variant is associated with multiple altered blood traits.
More specifically, the variant was associated with a decreased monocyte count and percentage, a decreased eosinophil count and percentage, and an increased neutrophil percentage.
The PGS analysis for potential target genes of the variant yielded consistent and robust evidence of pathways involving monocyte and eosinophil blood counts and weaker evidence of a pathway involving neutrophil counts.
Evidence also suggested basophils as another candidate pathway: the risk allele downregulates the expression of CCR3, reduces its stimulation of basophil production, and therefore lowers the basophil count.
The cell-type-specificity of their expression patterns also demonstrated the potential significance of these candidate genes. CCR3 was highly and specifically expressed in eosinophils and basophils, but only slightly expressed in neutrophils; CCR2 was expressed at high levels in basophils and moderate levels in monocytes, while CCR1 was expressed at medium to high levels across all types of granulocytes and monocytes.
The downstream pathophysiology involves hematologic processes
The team says their study has prioritized hematologic processes as the downstream pathophysiology of a significant genetic locus of severe COVID-19.
“Our phenome-wide association study for the severe COVID-19 risk variant at locus 3p21.31 and its candidate target genes identified altered blood cell traits, especially counts of monocyte, eosinophil, and neutrophil, as the probable mechanistic links between the genetic locus and severe COVID-19,” write the researchers.
“These blood cell traits, together with their candidate acting genes, CCR1, CCR2, and CCR3, represent compelling and testable hypotheses that call for follow-up studies into their roles in COVID-19 pathogenesis,” they conclude.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources