The COVID-19 pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has become a severe threat to global health. Research shows that the SARS-CoV-2 virus propagates by packaging its RNA genome into host cell membrane enclosures. Although it is known that the packing of the viral RNA into the nascent virion is mediated by the nucleocapsid (N) protein, the underlying mechanism of this is not clear.
Now, a study by researchers from the University of California Berkeley and the University of California San Francisco has shown that the N protein forms biomolecular condensates with the viral RNA in both in vitro and in vivo studies. The research titled, ‘SARS CoV-2 nucleocapsid protein forms condensates with viral genomic RNA’, was published on the preprint server bioRxiv* on September 14th, 2020.
The majority of methods developed so far to treat COVID-19 target the interactions between SARS-CoV-2 spike protein and the human ACE2 receptor and, thus, the initial stage of infection in the host cells. Not many studies focus on stopping virus proliferation in the host cells post-infection.
In order to propagate in the host cells, the viral genome first replicates, and the genomic RNA, bound by viral N protein, is then exported to the cytosol with the help of a molecular pore complex. The viral RNA and the N protein form viral ribonucleoprotein complexes (vRNPs), asymmetric “beads on a string” structures that bind to the viral membrane (M) protein. This triggers the budding of the vRNP complex and release of the enveloped virus into the extracellular space.
The SARS-CoV-2 N protein
According to some previous studies, the domain organization of the SARS-CoV N protein and its interaction with phase-separated structures show that the N protein phase separates with the viral genome in infected host cells.
The N protein has several of the characteristic features of phase separating systems, including an RNA binding domain (RBD), an uneven electrostatic charge distribution, intrinsically disordered regions, and a dimerization domain. The N protein also has a serine/arginine-rich (SR) motif, which helps with phase separation in other ribonucleoproteins, and a prion-like domain that can trigger protein demixing.
Phase separation of N protein can form membrane-less compartments that drive viral genome packaging in the host cells. Phase separation creates membrane-less organelles such as the nucleolus, heterochromatin, centrosome, stress granules, and P granules.
The study and its findings
In this study, the team of researchers expressed the N protein of SARS-CoV-2 virus in human embryonic kidney (HEK 293) cells to see if it could form a biomolecular condensate with viral genomic RNA in vitro. They observed that N protein and genomic RNA formed structures similar to the “beads on a string” pattern seen in infected host cells.
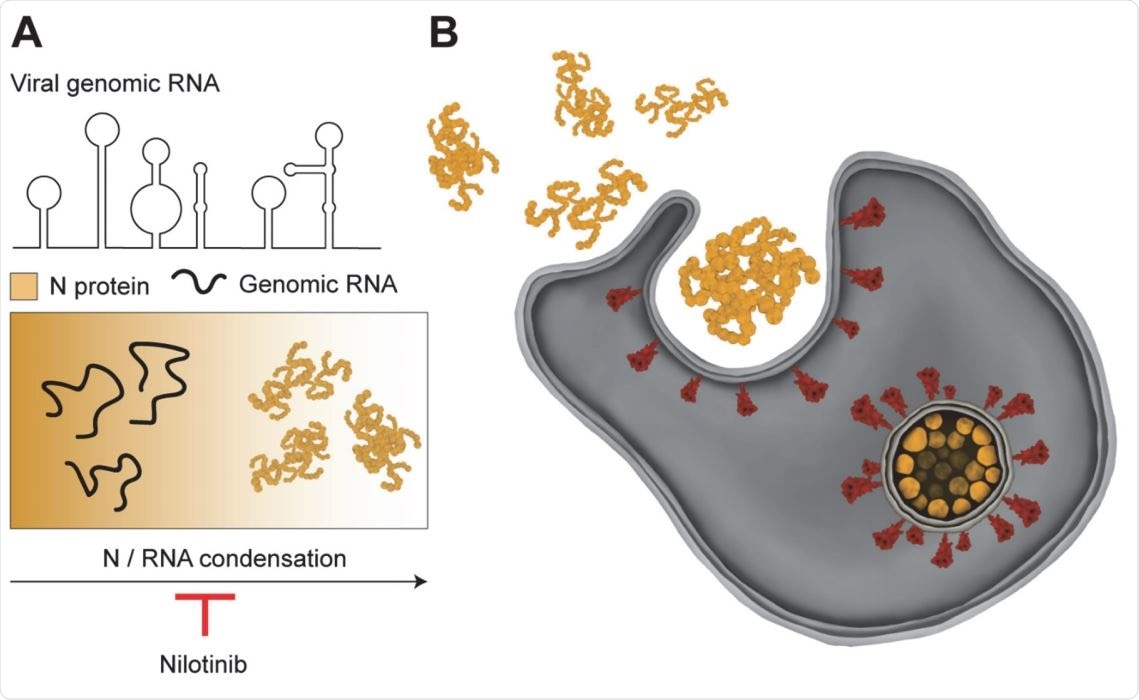
Model for remodeling of viral RNA genome by the SARS CoV-2 N protein. (A) The N protein packages viral genomic RNA into asymmetric gel-like structures through phase separation. Nilotinib disfavors formation of higher order structures by the N protein, and may disrupt packaging of the RNA genome into the viral envelope. (B) vRNPs interact with the viral membrane proteins on the surface of the ER-Golgi intermediate compartment and form the enveloped virus.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Using cross-linking mass spectrometry, the team was able to identify 2 regions flanking the dimerization domain. They found that the removal of the C-terminal oligomerization sequence completely disrupted phase separation.
“We observed that N and viral RNA form higher-order structures that resemble the “beads on a string” pattern observed in infected cells.”, says the team.
The highly phosphorylated SR motif in human cells promote host protein interactions, viral genome transcription, and nuclear targeting. Although previous reports suggest SR phosphorylation making N condensates more liquid in SARS-CoV-2, this study did not find any evidence of the SR motif driving phase separation of SARS-CoV-2 N protein in human cells.
On testing a library of FDA-approved molecules, the researchers identified drugs that can manipulate the size, shape, or number of the N protein droplets. One particular kinase inhibitor, nilotinib, showed promising ability to disrupt the in vitro condensation of N protein with viral RNA.
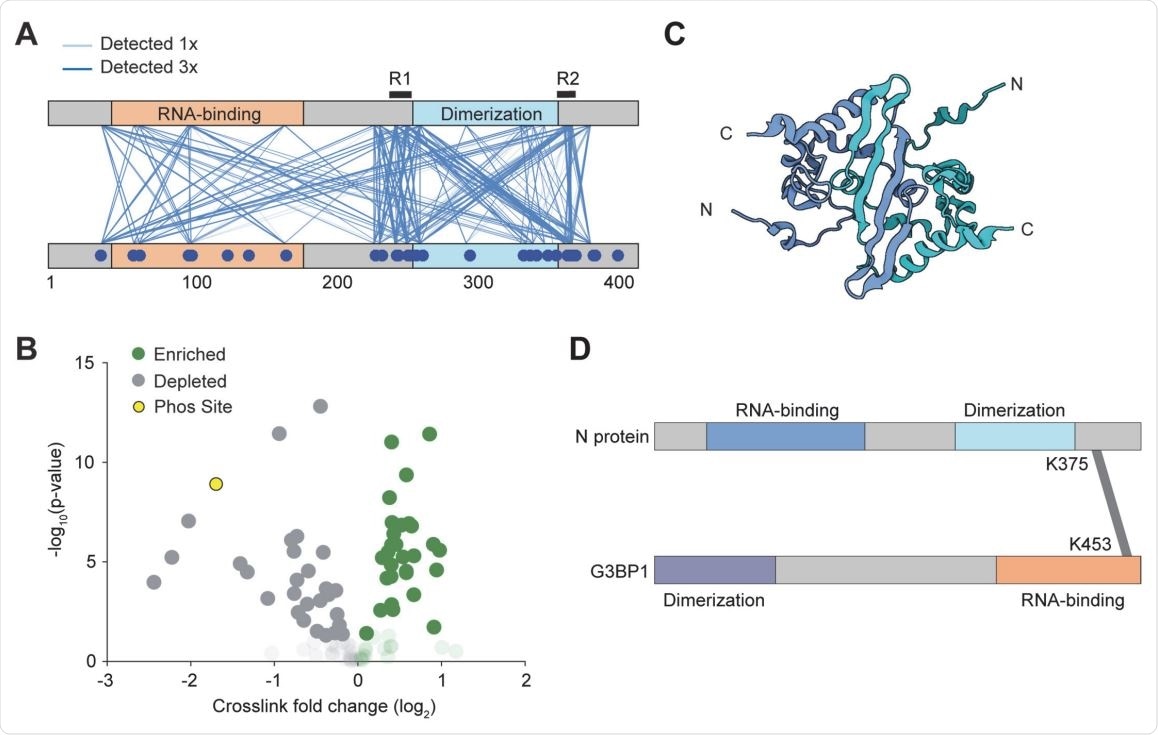
Interactions between N proteins and between N and G3BP1. (A) In an initial qualitative experiment, pairwise interactions were detected by CLMS at 300 mM salt. Lines depict a unique crosslink detected. The regions of N protein interactions flank the dimerization domain. (B) Volcano plot of the quantitative CLMS data comparing the droplet and no droplet condition. Opaque data points have a pvalue below 0.05, transparent data points have p-value greater than 0.05. Green represents unique crosslinks that are enriched in the droplet condition. The yellow markers represent the K169-K65 and K169(phosS176)- K65 crosslinks. (C) The structure of the dimerization domain (pdb# 2JW8 (64)). Because the N and C termini of the protomers are positioned away from each other, R1 and R2 within the same dimer are unlikely to interact with each other. (D) Mass spectrometry identified that the RNA binding domain of the stress granule protein G3BP1 interacts with the R2 region of N.
How the findings might help fight COVID-19?
By expressing the N protein in a mammalian system in vitro, the study showed that the N protein forms liquid condensates with unstructured RNA fragments and can also form a biomolecular condensate with viral RNA, which was consistent with previous studies.
Thanks to the fundamental role of the N protein in the viral genome packaging, researchers think that it could be a potential target of drugs used to treat COVID 19. This study provides valuable insights into how existing drugs used in the management of COVID-19 modulate the viral infection. One specific drug identified by the team, nilotinib, impacted phase separation with specific viral RNA and non-specific Poly-C RNA in vitro.
Although previous studies suggested nilotinib can inhibit the propagation of SARS-CoV and SARS-CoV-2, the mechanism of inhibition was unclear. The findings of this study clearly indicated that nilotinib inhibits the N/RNA condensate formation, which could disrupt viral replication.
“Our results indicate that nilotinib affects the formation of the N/RNA condensates, which could potentially impair viral replication.”, says the team
These results indicate the need for more such studies focusing on the role of nilotinib in treating
COVID-19 patients. Many COVID-19 clinical trials involving the parent drug of nilotinib, imatinib, are currently underway.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
SARS CoV-2 nucleocapsid protein forms condensates with viral genomic RNA Amanda Jack, Luke S Ferro, https://www.biorxiv.org/content/10.1101/2020.09.14.295824v2
- Peer reviewed and published scientific report.
Jack, Amanda, Luke S. Ferro, Michael J. Trnka, Eddie Wehri, Amrut Nadgir, Xammy Nguyenla, Douglas Fox, et al. 2021. “SARS-CoV-2 Nucleocapsid Protein Forms Condensates with Viral Genomic RNA.” Edited by Andrea Cimarelli. PLOS Biology 19 (10): e3001425. https://doi.org/10.1371/journal.pbio.3001425. https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3001425.