The rapid spread of deadly SARS-CoV-2 has put a massive burden on global public health. The human to human transmission and infectivity of SARS-CoV-2 primarily depends on the interaction between the receptor-binding domain (RBD) of viral spike protein and ACE2, a receptor expressed on the epithelial cells that line the human respiratory tract. Therefore, any research deciphering the variability of spike-ACE2 interaction across the human population is of prime importance in terms of developing appropriate therapeutics or vaccines to contain the progression of the COVID-19 pandemic.
Current study design
In the current study, the scientists tried to determine whether the SARS-CoV-2 spike protein recognizes the single-nucleotide variants of the human ACE2 receptor differentially. To address this question, they conducted deep mutational scanning to evaluate how more than 3,500 amino acid substitutions in the extracellular peptidase domain of the ACE2 receptor may influence the interaction with the viral spike protein. From the scanning data, they found many novel ACE2 residues that impact the spike binding pattern and identified more than 100 single-nucleotide variants of ACE2 that are likely to be differentially recognized by the viral protein.
Important observations
Precisely, the scientists identified 597 locations in the ACE2’s extracellular domain containing 3,571 amino acid substitutions. Of these substitutions, 68% were associated with reduced spike binding efficiency, and 4% were associated with increased spike binding efficiency. About 28% of the substitution had no statistically significant effect on the spike-ACE2 interaction. Interestingly, they observed that these amino acid substitutions included 84% of all ACE2 missense single-nucleotide variants identified in the human population. A total of 165 ACE2 variants were identified in the current study. The majority of these variants were distant from the spike-ACE2 interface, indicating that the spike-ACE2 interaction can be modulated by ACE2 mutations that are distal to the spike binding interface.
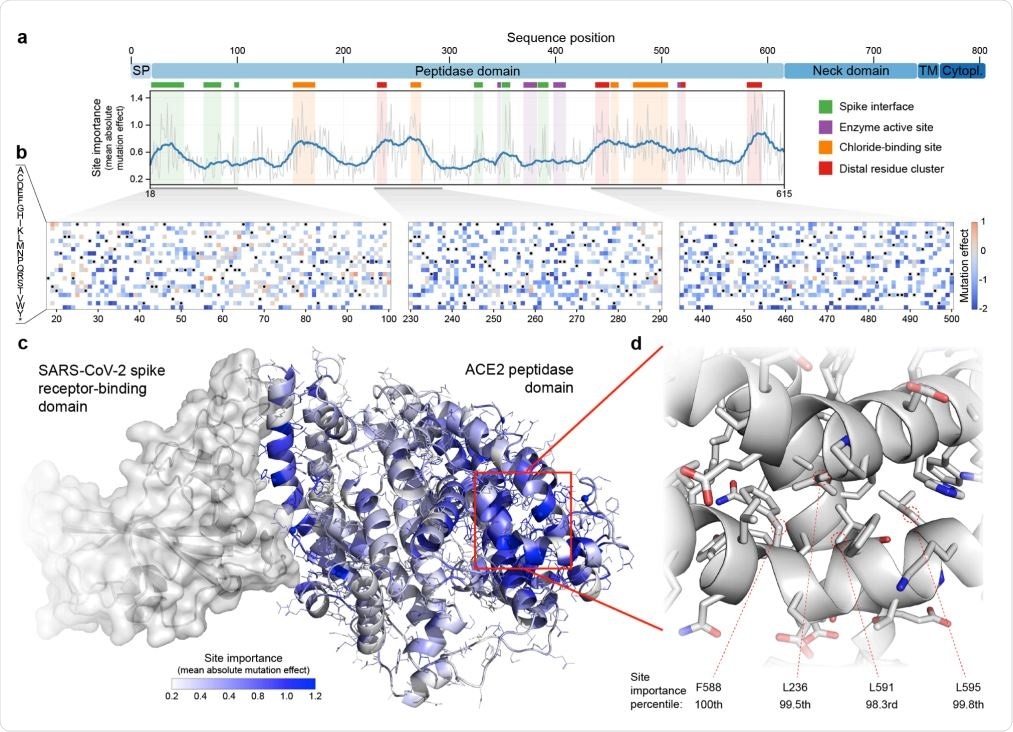
Large-scale mutagenesis of ACE2’s peptidase domain. (a) Analysis of how amino acid substitutions across positions 18-615 affect binding of the SARS-CoV-2 spike protein. The plot quantifies the importance of each site by taking the mean of the absolute value of all mutation effects observed at that site. The grey line represents the mean absolute value of the mutation effect and the blue line shows the moving average to highlight general regions of ACE2 that are important for binding. Key structural landmarks are highlighted with shaded regions along the length of the sequence. (b) Mutation effect heat maps for three different regions of ACE2. Red denotes mutations that increase ACE2 spike binding; blue denotes reduced binding. Overall, we observe the effects of 3571 amino acid substitutions across 597 positions in ACE2’s peptidase domain. (c) The mean absolute mutation effect mapped onto the threedimensional ACE2 structure (PDB ID: 6LZG). Residues near the spike interface are important for binding, in addition to many sites located on the distal lobe of the protein domain. (d) The most important region of ACE2 structure is composed of a tightly packed cluster of residues located over 30 Å from the spike interface.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
To validate the accuracy of current study data, the scientists evaluated the binding impact of previously identified important ACE2 residues (S19, Q24, D30, H34, D38, Y41, Q42, Y83, and K353) that are present at the interface between the viral spike protein and the α-helical and β-turn sequences of ACE2. In agreement with previous study findings, they observed that these residues play essential roles in mediating the spike-ACE2 interaction.
The novelty of the study is the identification of previously unrecognized ACE2 residues that are crucial for spike binding. Precisely, the scientists observed that some residues adjacent to the chloride-binding domain of ACE2 play vital roles in modulating the spike-ACE2 interaction by altering the structural conformation of ACE2.
By analyzing the allele frequency of each ACE2 variant in the general population, the scientist estimated that about 320 to 365 of 100,000 individuals carry ACE2 variants that may reduce the spike binding; whereas, 4 to 12 of 100,000 individuals carry ACE2 variants that may increase the spike binding. Interestingly, they observed that the frequency of having specific variants that alter the spike binding varies between people with a different ancestral origin. For example, compared to the general population, the African population is five times more likely to carry specific ACE2 variants, such as p.Met82Ile.
Regarding the mode of action of ACE2 variants, the scientists revealed that the variants with reduced binding efficiency have a three-fold reduction in maximum binding signals than that of wild-type ACE2. Moreover, the ACE2 variants identified in the study were significantly different from wild-type ACE2 in terms of spike binding affinity, the number of receptors present on the cell membrane, and receptor turnover rate at the cell membrane.
Collectively, the study findings provide significant insight into basic SARS-CoV-2 research and pave the way toward the identification of novel therapeutic targets.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources