A part of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral RNA is highly conserved. Researchers have used this as a target for designing antisense oligonucleotide gapmers to cleave the RNA and disrupt virus replication. Their research is published as a preprint on the online server bioRxiv* in September 2020.
The spread of COVID-19 caused by the SARS-CoV-2 has led to the development of different methods of combating the pandemic. Several types of vaccines are in various stages of trials. Other studies have investigated if drugs approved for other diseases (repurposed drugs) might be useful as a treatment for COVID-19 disease. Another approach is to develop new methods to disrupt SARS-CoV-2 viral replication.
The SARS-CoV-2 virus is a single-stranded RNA virus. The virus replicates inside the host cell using enzymes, which are encoded by the virus. The viral genome also has many structural elements whose functions remain largely unknown.
One such element is the stem-loop 2 motif (s2m), a highly conserved sequence in the Sarbecovirus genus, which includes SARS-CoV and SARS-CoV-2. The s2m is nearly identical in these two viruses, with only a 2-nucleotide difference and is always present in the samples taken from patients who have tested positive for SARS-CoV-2.
Because s2m is highly conserved, with a low probability of it mutating, researchers believe it to be a promising target for developing antivirals.
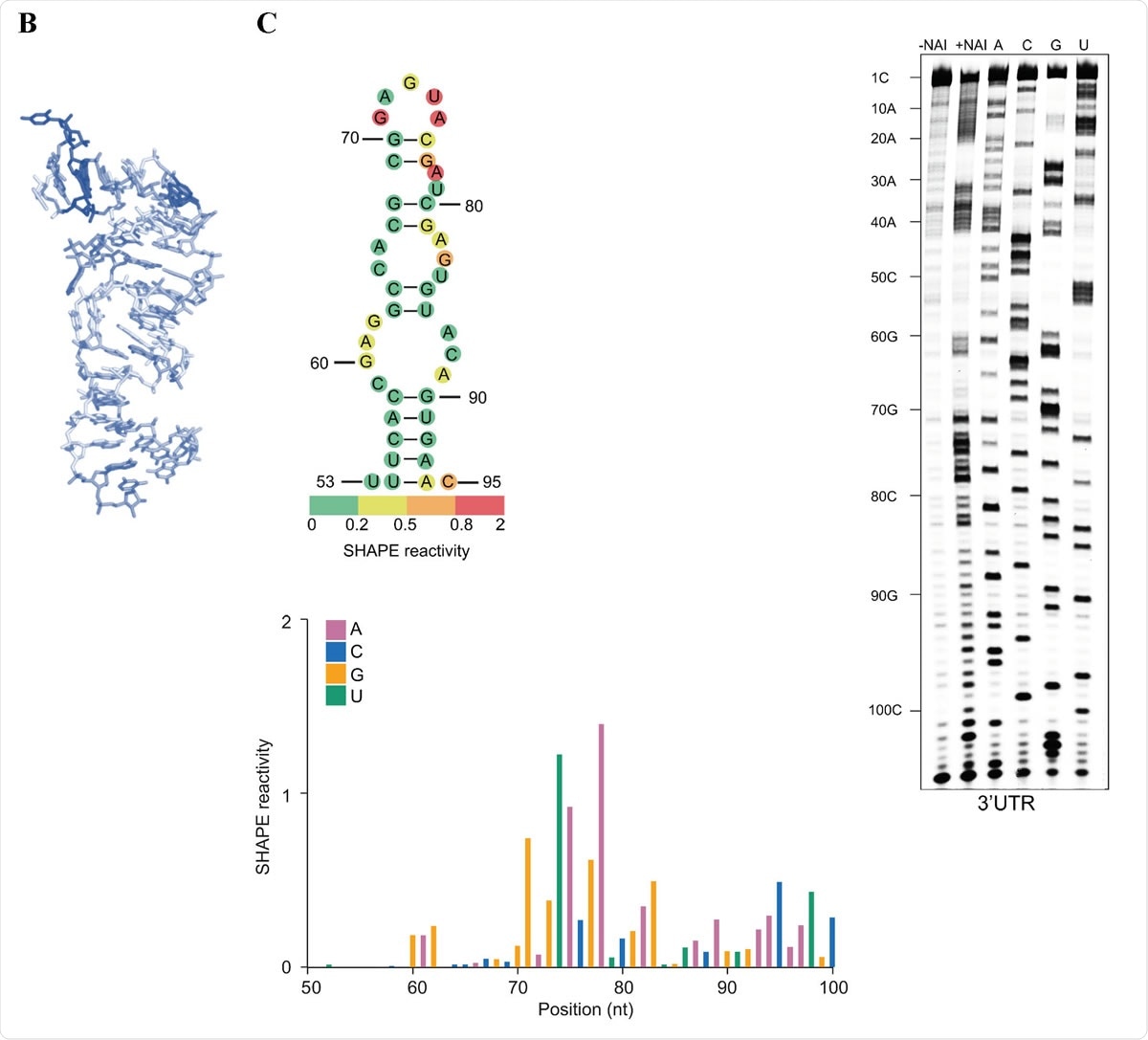
s2m is a conserved structural element in the SARS-CoV-2 genome. (A) Sequence alignment of the s2m element in the 3ʹ UTRs of SARS-CoV-2 and SARS-CoV. (B) The crystal structure of the SARS-CoV s2m element (adapted from Robertson et al., 2005). (C) Chemical probing of the 3ʹ UTR of SARS-CoV-2. RNA was denatured and refolded in the presence of 100 mM K+ and 0.5 mM Mg2+, then incubated with NAI (+NAI channel) or DMSO control (-NAI channel). NAI modification was detected by reverse transcription stalling and gel-based analysis. Sequencing lanes were generated by adding ddT (for A), ddG (for C), ddC (for G) and ddA (for U) when performing reverse transcription. The lower panel shows quantification of SHAPE signal in the s2m and flanking regions. Calculation was based on the gel in Fig. 1C, by subtracting the signal of the +NAI lane from that of the -NAI lane. The upper panel shows annotation of SHAPE signal on the s2m structure. The bases with SHAPE signal of 0-0.2, 0.2-0.5, 0.5-0.8 and 0.8-2 were coloured with green, yellow, orange and red, respectively.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Designing gapmers and in vitro testing
In the new study, an international team of researchers used locked nucleic acid (LNA) antisense oligonucleotides (ASO) to target the s2m element. ASOs are a sequence of single-stranded deoxyribonucleotides complementary to the target. LNA is a type of ASO where the conformational freedom of the nucleotides is “locked.” One version of these types of LNA ASO is the gapmer, where LNA bases are present at the edges of a central DNA sequence. The central DNA then uses ribonuclease H (RNase H) to cleave RNA, while the gapmer remains intact and can bind to other RNA molecules.
First, the team investigated the structure of the s2m element, and concluded that the structure folds into a stem-loop structure.
Since the target structure is folded, they designed gapmer LNA bases to bind to the exposed bases of s2m, which after initial binding, can lead to the unfolding of s2m as the rest of the gapmer to binds to the complementary nucleotides. Gapmers 1-3 were designed to have a higher affinity for the target RNA than gapmers 4-6.
Testing the designed gapmers in vitro, the team found that all the 6 gapmers degraded s2m RNA, even when twice the number of RNA molecules for every gapmer molecule, indicating the gapmers were reused.
They also looked at the structure of the s2m element in the presence of the gapmers using a technique that determines how accessible are the nucleotides. When gapmers 1 and 2 were present, intermolecular interactions increased between the gapmers and the target sequences, and these regions became more like single strands. “Thus, the inter-molecular interactions between gapmers and target regions could also remodel the folding status of flanking regions,” write the authors.
Gapmers inhibit replication in human cells
For testing the effect of the gapmers in human cells, the researchers used HeLa and A549 reporter cells derived from the lungs with green fluorescent proteins (GFP).
In the presence of the gapmers 1-3 and gapmers 5-6, GFP fluorescence decreased, indicating inhibition of replication. Gapmer 4, which had only six nucleotides, showed no effect, suggesting it may be too short to recruit human RNase H in vivo.
Astroviruses are another type of single-stranded RNA viruses that also have the s2m structure. The team had previously developed a human astrovirus 1 (HAstV1)-based replicon system, which they now modified to include the SARS-CoV-2 and SARS-CoV s2m element, to test their gapmers. Since the astrovirus model system has a smaller genome size than the coronavirus, it is easier to study.
Gapmers 1, 2, and 5 effectively inhibited the replication of replicons with SARS-CoV-2 s2m structure, at sub-nanomolar concentrations. Inhibition of replication using gapmers 3, 4, and 6 was less pronounced.
A point to note, write the authors, is that the SARS-CoV-2 viral RNA is concealed in membrane-like materials in the host cell. This may prevent ASOs from reaching the s2m element of the virus. Hence, gapmers that improve association with cell membranes may help them reach the RNA. LNA ASOs have been conjugated with tocopherol and cholesterol before to help membrane binding. Such designs may help membrane association, helping ASOs to reach the RNA.
The authors say these results are promising to continue testing gapmer and other ASO-based strategies for inhibiting RNA replication in SARS-CoV-2. However, further testing and optimizing will be required to make effective therapeutics, including testing in cell cultures and animal models.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources