By using a modern genome-wide knockout approach to single out coronavirus host factors in human cells, researchers from Rega Institute in Belgium discovered a potential new target that might be exploited for developing drugs against the ongoing coronavirus disease (COVID-19) pandemic, but also future outbreaks of highly pathogenic coronaviruses. The paper is currently available on the bioRxiv* preprint server.
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which instigated the COVID-19 pandemic, is responsible for global economic damage and one million deaths. Thus far, the quest for an effective drug or vaccine has been elusive.
To date, there are seven known human coronaviruses, which likely all emerged as a zoonosis from bats, mice, or other animals. Four of them are so-called 'common cold human coronaviruses' that cause only mild upper respiratory tract illness: alphacoronaviruses 229E and NL63, and betacoronaviruses OC43 and HKU1.
Conversely, the betacoronaviruses SARS-CoV, MERS-CoV, and the most recent SARS-CoV-2 are highly pathogenic and responsible for severe and potentially lethal respiratory tract infections. Since animals harbor a broad diversity of coronavirus types with known interspecies transmission, there is a high likelihood that a new (potentially pandemic) strain will emerge in the future.
But despite this risk, our options to either prevent or treat any coronavirus infection are very limited. Therefore, the development of broad-spectrum antivirals against the members of this virus family is not only necessary for this pandemic, but also to swiftly tackle and contain similar zoonotic events in the future.
In an exciting new research endeavor, scientists from Rega Institute in Leuven (Belgium) conducted a series of genome-wide CRISPR-based genetic screens in human cells in order to elucidate host factors that are needed for SARS-CoV-2 and HCoV-229E infection, and propose potential therapeutic solutions.
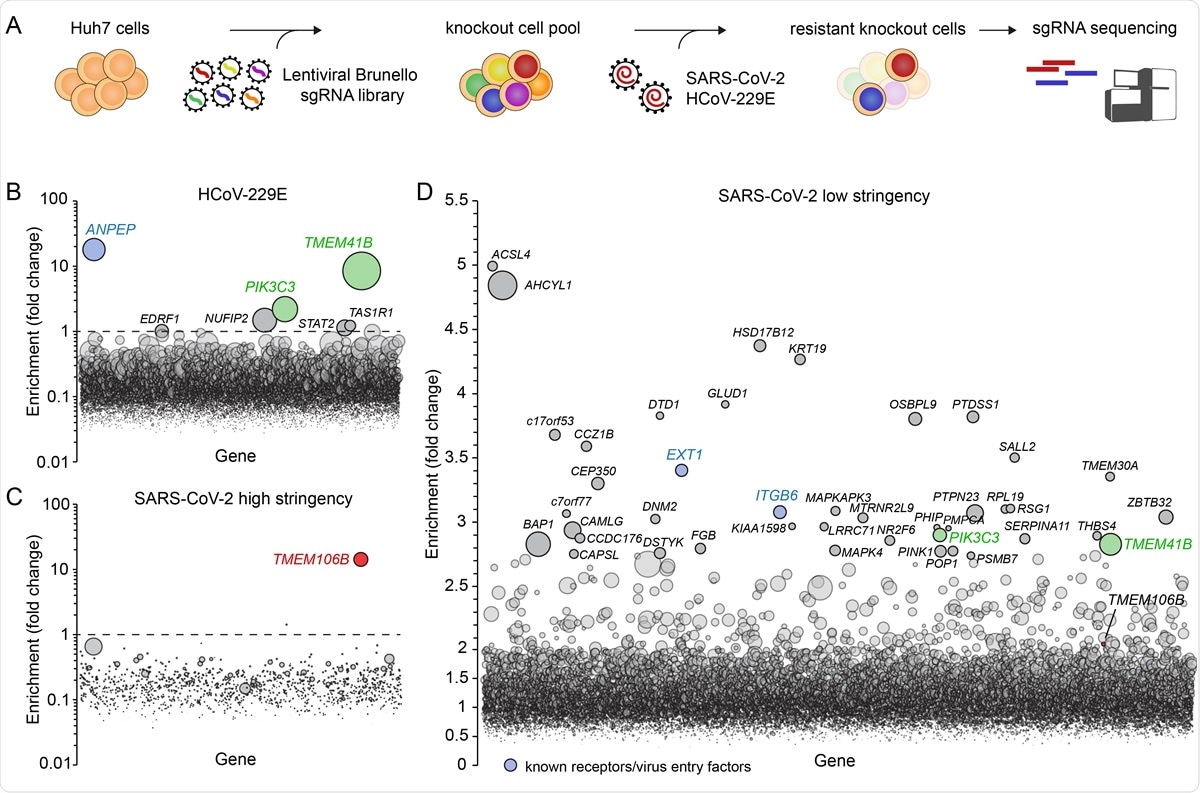
Genome-wide knockout screens in human cells identify 105 host factors for SARS-CoV-2 and HCoV-229E infection. A) Overview of experimental steps performed during a genome-wide screen for coronavirus host factors. B-D) Genome-wide knockout screens were performed in Huh7 cells, with strong selection (high stringency) using HCoV-229E (B) and SARS-CoV-2 (C) or with mild SARS-CoV-2 selection (low stringency) (D). Each circle represents a gene, with size corresponding to significance of enrichment. The y-axis shows the enrichment of sgRNAs after virus selection compared to an uninfected control population (D) or the population on the first day of the screen prior to infection (B and C). Genes distributed on the x-axis in alphabetical order.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
A genome-wide knockout screen
In this study, the aforementioned genome-wide knockout screen was performed in human cells without introducing an exogenous receptor. To that end, the researchers have used the human liver carcinoma cell line Huh7, where a clinical isolate of SARS-CoV-2 induced a straightforward cytopathic effect naturally. Less pathogenic HCoV-229E was used as a control strain.
This allowed the researchers to identify both broad-spectrum coronavirus host factors, as well as specific host factors for SARS-CoV-2 and HCoV-229E. Huh7 cells were transduced with the Brunello genome-wide library, which was followed by single-guide RNA (sgRNA) identification by deep sequencing.
Several pharmacological inhibitors were used to confirm the role of class III phosphoinositide 3-kinase (PI3K in coronavirus infection, which is known to play a role in many processes – including endocytic trafficking and autophagosome initiation and maturation.
Common and specific cellular host factors
"We identified PI3K type 3 as a common host factor for SARS-CoV-2, HCoV-229E, and HCoV-OC43, and show that small molecules targeting this protein might serve as broadly applicable anti-coronavirus inhibitors", study authors explain their findings.
Their data show that PI3K type 3 plays a substantial role in early steps of the SARS-CoV-2 life cycle, such as endocytosis, fusion, translation or early onset of replication. Likewise, this kinase supports infection by inducing phagophore nucleation or autophagosome precursor membrane formation.
The researchers have also demonstrated that TMEM41B – the important regulator of autophagy (i.e., our body's way of removing damaged cells in order to regenerate new ones) – is indispensable for HCoV-229E infection, and pinpointed TMEM106B as a new cellular host factor crucial for SARS-CoV-2 infection.
More specifically, TMEM106B may promote acidification of vesicles in the endolysosomal pathway, facilitating the efficient delivery of the SARS-CoV-2 genome into the cytoplasm of human cells, playing a role early in the viral replication cycle (i.e., in the first six hours after infection).
On the other hand, in their SARS-CoV-2 screens, the researchers did not identify ACE2, which is in stark contrast to previous screens that were performed in Vero E6 cells or engineered human cells that overexpress ACE2. Furthermore, it was shown that heparan sulfate is rather important for efficient infection.
A viable option for today and the future
"Our results identify new coronavirus host factors that may potentially serve as drug targets against SARS-CoV-2 or to quickly combat future zoonotic coronavirus outbreaks", study authors summarize their findings in bioRxiv preprint paper.
The periodic emergence of new pathogenic coronaviruses indeed poses a serious public health threat beyond the current COVID-19 pandemic, which means pan-coronavirus inhibitors (like the one presented in this paper) are very much needed in our treatment armamentarium.
Nonetheless, this study also tackles the notion that different, albeit related, viral agents may depend on distinct factors to exploit similar cellular pathways. Hence, all possibilities of developing a broad-spectrum viral inhibitor (particularly for deadly viruses) should be embraced and further researched.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources