Derek Corcoran and colleagues found that models of early COVID-19 growth dynamics that included the different clades the virus has mutated into significantly improved growth rate predictions.
Including clades in the models was also more important than incorporating the SARS-CoV-2 variant D614G, which has been associated with increased viral load and infectivity.
More specifically, a higher prevalence of the clades 19A and 19B, which emerged during the Wuhan outbreak, correlated with lower growth rates. Higher prevalence of the clades 20A and 20C, which emerged from 19A and was prominent early on in the European outbreak, correlated with higher growth rates.
The researchers say that without intervention, COVID-19 has the potential to grow more quickly in regions dominated by the 20A and 20C clades, which includes most of South and North America.
A pre-print version of the paper is available on the server medRxiv*, while the article undergoes peer review.
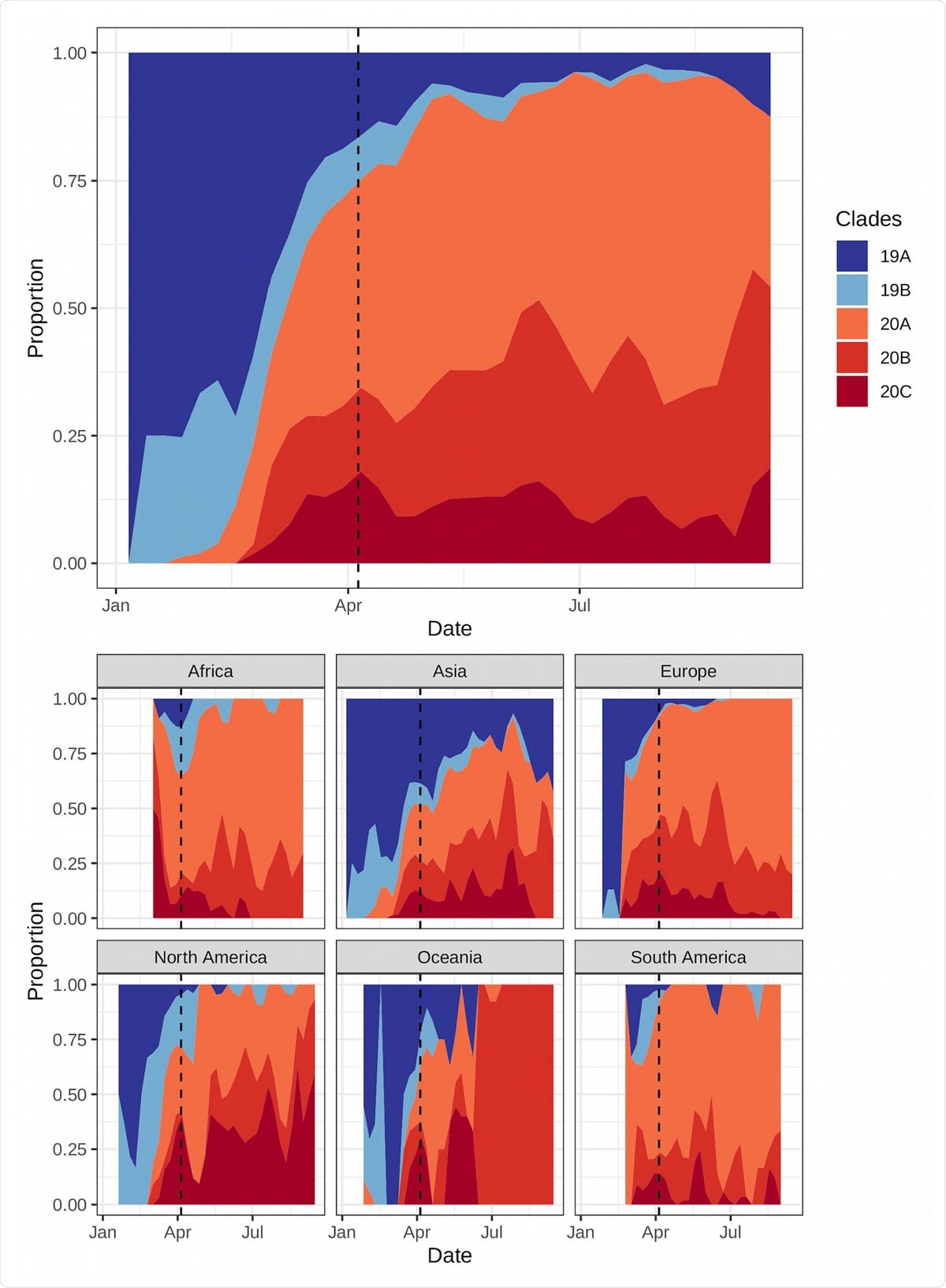
Proportions of each clade globally (top) and by region (bottom). Clade 19 (blues) diminished over time, while clade 20 (reds) increased during that same period. Among the type 20 clades, 20A and 20B have increased to similar proportions globally while 20C had more moderate proportional growth. However, clade 20C has increased most dramatically in South and North America, two regions where COVID-19 has subsequently grown quickly. The dashed line represents the last date used for fitting the models.

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Models do not currently include different genetic clades
As SARS-CoV-2 has spread around across the globe infecting millions of people, it has mutated into divergent clades with different prevalence rates, depending on geographical region.
Although most of the mutations involved are expected to be inconsequential, some mutations in the SARS-CoV-2 spike protein have been proposed to affect the critical stage at which the virus infects human cells, which could alter disease transmission and dynamics.
“If true, then we expect an increased growth rate of reported COVID-19 cases in regions dominated by viruses with these altered proteins,” say the researchers.
However, the models currently used to inform mitigation measures and healthcare capacity generally assume equivalent transmission probability and pathogenicity.
“Understanding if genetic clades differ in their infectivity and, if so, where they are most prevalent would aid efforts to design effective intervention strategies that control the virus and end the pandemic,” writes the team.
Although some clinical studies have shown the SARS-CoV-2 variant D614G is associated with increased viral load, it is unclear whether carriers of this mutation are more infectious, and there is no evidence to suggest the variant is associated with increased virulence.
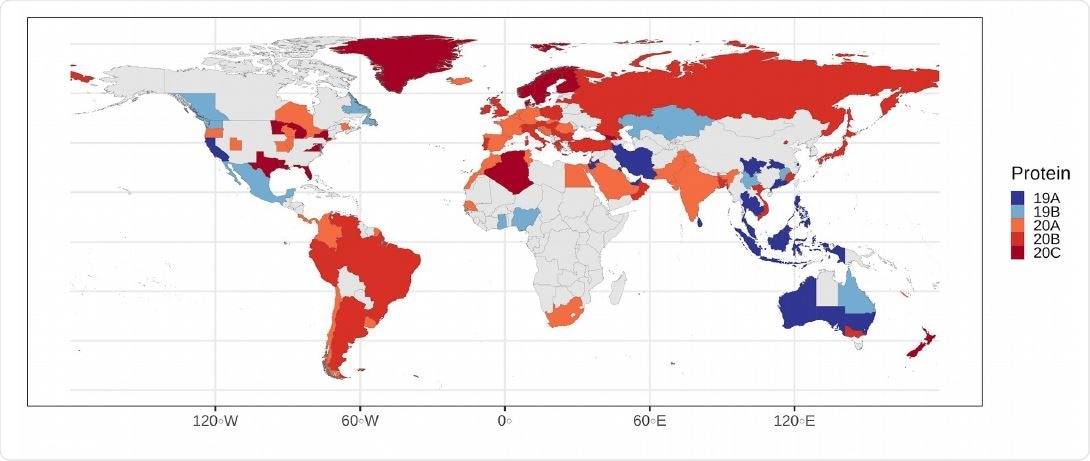
Most common clade on each polity as seen in the map, clades 19A and 19B dominate in Oceania and southeast Asia, whereas clades 20A, 20B and 20C dominate in South America, Europe and North America.
What did the researchers do?
Corcoran and colleagues tested whether the prevalence of certain clades or the D614G variant within a population was associated with a higher growth rate of the disease.
The team-based their model on a previous model of early infection dynamics that highlighted the effects of other potentially important factors, including ultraviolet light, temperature, humidity, and the age structure of populations.
What did the researchers find?
The researchers found no evidence that variant D164G alone contributed to the early growth rate of COVID-19.
Clade assignment, on the other hand, did enhance the ability to describe COVID-19 growth dynamics early on in the pandemic. Including clade in the models significantly improved predictions compared with previous studies that only included weather and demographic variables.
Higher proportions of clades 20A and 20C were associated with higher rates of COVID-19 growth, while higher proportions of clades 19 A and 19B were associated with lower growth rates.
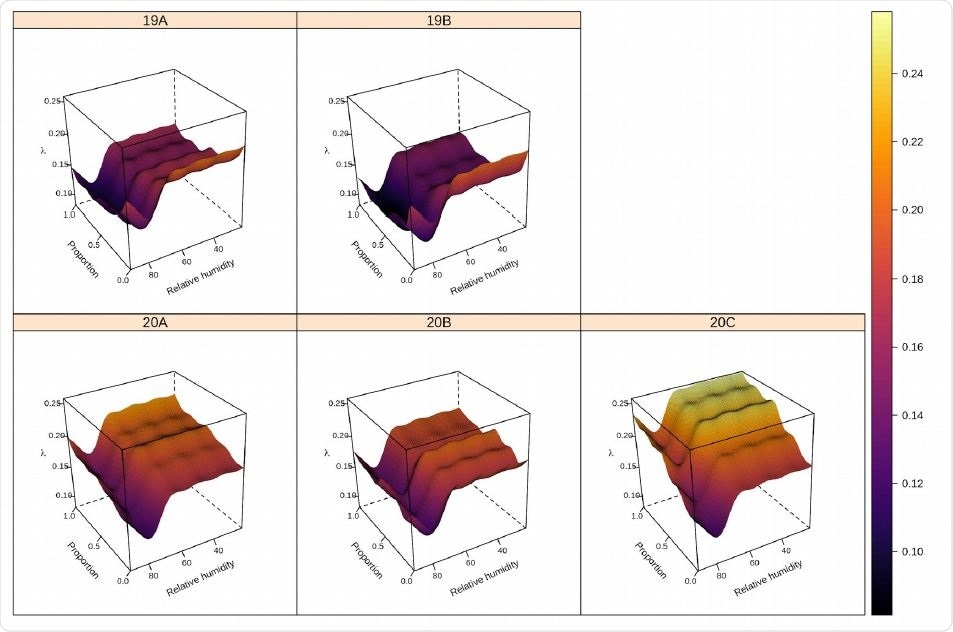
Predicted COVID-19 growth rate (color gradient from dark blue to yellow) based on the interaction between relative humidity and the proportion of clade 20C. Higher proportions of clade 20C and lower relative humidity are associated with higher values of COVID-19 growth rate. The clumps are a byproduct of GAMsmoothing across the discrete nature of classifications by nodes in Boosted Regression Trees.
Humidity seemed to play a role
The team also found that humidity seemed to interact with viral clades in a way that may increase or decrease infectivity.
For example, clades 20A and 20C generated the highest growth rates when they were coupled with low humidity.
The researchers say this suggests that the impact of clade identity might be more important when coupled with certain weather conditions.
Growth rates might increase in Africa, South America, and North America
The team also reports that during August, Africa, South America, and North America had the highest proportion of clades 20A and 20C, suggesting the potential for increased COVID-19 growth rates in these three continental areas in the future.
Corcoran and colleagues point out that although a correlation between COVID-19 growth rate and the prevalence of certain clades was identified, no conclusions can yet be drawn about causality. Causality and correlation could only be separated out through controlled trials.
In the meantime, continued genomic monitoring is critical to detect whether certain clades associated with the current pandemic or future ones change in frequency over time, say the researchers.
“As we understand more about the evolution of disease agents, such as SARS-Cov-2, this information should aid our ability to make accurate predictions about future outbreaks and design effective public health interventions in order to end this pandemic and prevent new ones from emerging,” they conclude.

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.