Researchers in the UK have conducted a large-scale whole-genome sequencing study of clinical samples positive for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the Norfolk area to shed light on the origins, genetic variation, transmission, and spread of the virus in the region.
SARS-CoV-2 is the agent responsible for the current coronavirus 2019 (COVID-19) pandemic that is increasingly posing a threat to public health and the economy worldwide.
In collaboration with the COVID-19 Genomics UK consortium, researchers at Norwich Research Park and various other institutions in the UK analyzed genome sequencing data for SARS-CoV-2-positive samples collected from four major hospitals and various minor hospitals, care facilities, and community organizations within Norfolk and surrounding areas.
The team’s in-depth assessment of the genetic epidemiology of SARS-CoV-2 in this region during the first wave of the pandemic (March to August 2020) provided actionable information for future management strategies.
Examples included identifying a specific viral lineage associated with six care facilities, the ruling out of a nosocomial outbreak, and confirmation of an outbreak in a food processing facility.
A pre-print version of the paper is available in the server medRxiv*, while the article undergoes peer review.
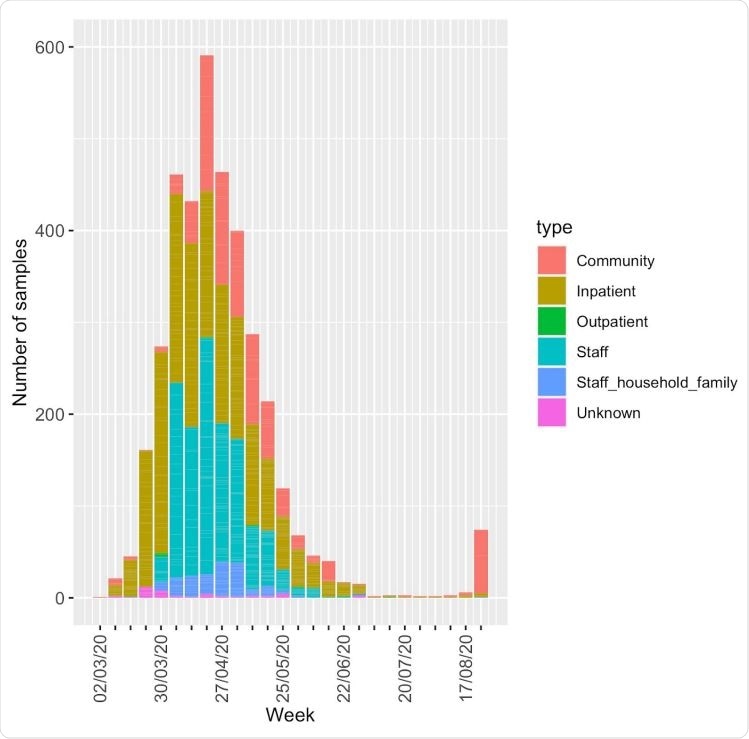
Total number of positive samples in the region per week, broken down by type. Not all of these were available for sequencing and a single individual may have been sampled multiple times. Staff (key workers) include healthcare workers and essential workers, such as police officers.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Understanding more about the evolution, transmission and spread of the virus
Since the first cases of COVID-19 were identified in Wuhan, China, late last year, the causative agent –SARS-CoV-2 – has spread to almost every country in the world.
In the UK, the initial peak occurred in April 2020, and in the county of Norfolk and surrounding areas, more than 3,200 cases were reported between March and August.
The COVID-19 Genomics UK (COG-UK) consortium is a nation-wide public health surveillance initiative that was set up to generate and study large-scale SARS-CoV-2 sequencing datasets, intending to understand more about the evolution, transmission, and spread of the virus in the UK.
What did the researchers do?
As part of the consortium’s activities, Andrew Page (Norwich Research Park) and colleagues performed whole genome sequencing of SARS-CoV-2 genomes present in clinical samples taken from four major hospitals, five smaller hospitals, three community care organizations and drive through testing facilities in Norfolk and surrounding areas.
Clinical metadata was combined with the genome sequencing data to investigate the origins, genetic variation, transmission, and spread of SARS-CoV-2 within the region of Norfolk and to add context to the national and global data.
“Weekly sequencing data were fed back into the national effort for pandemic management, while simultaneously being used to assist local outbreak analyses,” writes the team.
The researchers evaluated 1,565 SARS-CoV-2-positive samples from 1,376 cases, collected between March and August 2020. This represented 42.6% of all cases identified by hospital testing in the local population, including those with a clinical need, key workers, and their family members.
SARS-CoV-2 genomes from 1,035 of the cases were of a quality high enough to provide phylogenetic lineages.
What did the study find?
In the region, 26 distinct global lineages were observed, suggesting that genomically distinct lineages have expanded worldwide and indicating multiple separate introductions of the virus into the region.
Furthermore, 100 genetically-distinct UK lineages were observed, demonstrating local evolution, says the team.
As the number of infections cases increased during the course of the pandemic, more lineages were identified, with the number of co-occurring lineages peaking at around five to six weeks after a national lockdown was implemented.
“Thereafter, the number of lineages dropped substantially, with many rapidly becoming extinct in the region, providing further evidence that lockdown measures break transmission,” writes the team.
The team identified a sustained outbreak in care facilities
The researchers say their data unexpectedly identified a sustained outbreak in six care facilities.
While examining a common lineage within the region, the team noticed that positive samples had mainly come from older adults, suggesting a potential link to care facilities. Further investigation revealed that six care facilities shared a distinct sublineage that was primarily found only in those facilities.
The lineage data also ruled out a nosocomial outbreak in Ipswich Hospital. On sequencing 31 samples from the hospital, the researchers identified eight UK lineages, the most common of which was also the most commonly observed lineage in the UK.
“This demonstrated that a single large nosocomial outbreak had not occurred but that the patients in hospital had become infected in the community by circulating lineages,” said Page and team.
Transmission of a new lineage within a food processing facility
Finally, genome sequencing applied to a food processing facility identified virtually identical genomes that were not present in the general community before the outbreak, thereby demonstrating transmission of a new lineage within the factory.
“The factory sub-lineage is sufficiently novel to link it to the factory if identified anywhere in the country,” write the researchers.
Page and colleagues say the findings demonstrate the valuable role of large-scale genome sequencing of SARS-CoV-2 to inform surveillance and regional outbreak management.
“The large-scale genome sequencing of SARS-CoV-2-positive samples has provided valuable additional data for public health epidemiology in the Norfolk region, and will continue to help identify and untangle hidden transmission chains as the pandemic evolves,” concludes the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Page A, et al. Large scale sequencing of SARS-CoV-2 genomes from one region allows detailed epidemiology and enables local outbreak management. medRxiv, 2020. doi: https://doi.org/10.1101/2020.09.28.20201475
- Peer reviewed and published scientific report.
Page, Andrew J., Alison E. Mather, Thanh Le-Viet, Emma J. Meader, Nabil-Fareed Alikhan, Gemma L. Kay, Leonardo de Oliveira Martins, et al. 2021. “Large-Scale Sequencing of SARS-CoV-2 Genomes from One Region Allows Detailed Epidemiology and Enables Local Outbreak Management.” Microbial Genomics 7 (6). https://doi.org/10.1099/mgen.0.000589. https://www.microbiologyresearch.org/content/journal/mgen/10.1099/mgen.0.000589.