Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) first emerged in December 2019 in Wuhan, China, and rapidly spread to other countries, causing an unprecedented global pandemic.
Today, the virus has spread to 207 countries, with 35.65 million confirmed cases globally and over 1.04 million fatalities. Although it is known from previous experience with SARS-CoV-1 and the Middle East respiratory syndrome (MERS) that coronaviruses are highly virulent, there is still no FDA-approved vaccine or drug for any coronavirus.
Since SARS-CoV-2 is closely related to SARS-CoV-1 and MERS coronaviruses, it is classified as Risk Group 3 select agents, which means the use of the live viruses is restricted to biosafety level 3 (BSL-3) facilities. This makes SARS-CoV-2 research inaccessible to most functioning research laboratories in the US, inhibiting a collective scientific effort that such a pandemic calls for.
Creating BSL-2 models to facilitate viral research
The development of BSL-2 compatible viral models and assays is imperative to facilitate the study of viral budding and entry and potential therapeutic approaches. In the past, BSL-2 models of other highly virulent BSL-3 and -4 pathogens, including SARS-CoV-1, MERS, Ebola virus, and Lassa virus, have been developed in the form of virus-like particles (VLPs).
SARS-CoV-2, like other coronaviruses, uses angiotensin-converting enzyme 2 (ACE2), the human cell surface receptor, to enable cellular uptake of viral particles. The 4 structural proteins of the virus - spike (S), membrane (M), nucleocapsid (N), and envelope (E) - are said to be responsible for maintaining the structural integrity of the SARS-CoV-2 virion.
In a recent preprint paper published on the bioRxiv* server, researchers from the Purdue Institute of Inflammation, Immunology, and Infectious Disease, Purdue University, West Lafayette, IN, discuss their efforts to assess the 4 SARS-CoV-2 structural proteins for their ability to produce VLPs from human cells to create a competent system for BSL-2 studies of SARS-CoV-2. The study aimed to develop VLPs that are morphologically and functionally relevant that help study SARS-CoV-2 budding and entry.
In their work, the team of researchers provides methods and resources for production, purification, and fluorescent- and APEX2-labeling of SARS-CoV-2 VLPs, all of which can help evaluate viral budding and entry mechanisms and analysis of drug inhibitors under BSL-2 conditions.
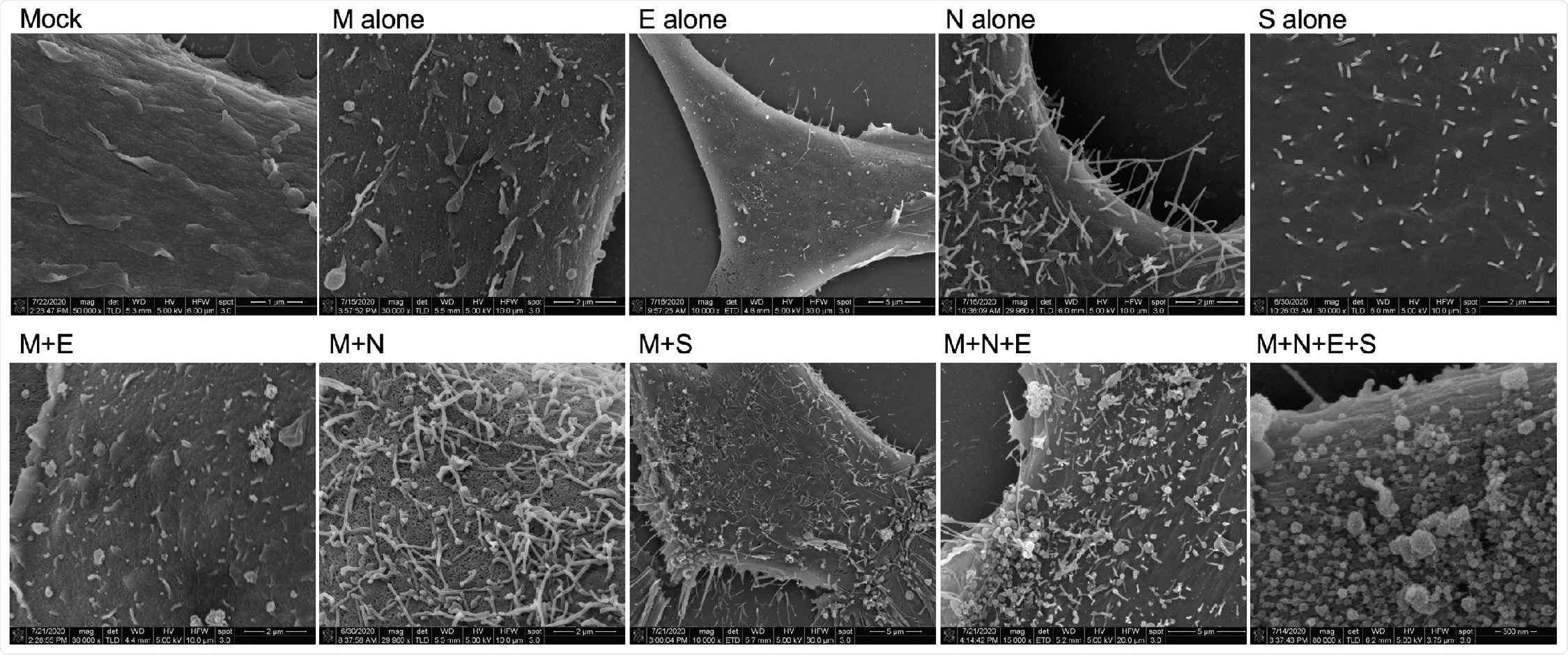
Scanning electron microscopy of viral structural protein transfected cells. HEK293 cells were seeded onto coverslips and transfected individually or in combination with M, N, E, and/or S. Cells were fixed with glutaraldehyde 72-hours post-transfection and kept at 4°C until fixed with osmium tetroxide. Samples were then gradually dehydrated with ethanol and completely dehydrated with a critical point dryer. Once dehydrated, samples were mounted onto aluminum pins with double-sided carbon tape, charged with silver paint, and sputter coated prior to imaging. Images range in magnification from 10,000x to 80,000x.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
A realistic model of SARS-CoV-2 viral entry available in BSL-2 setting
As the SARS-CoV-2 pandemic continues to rage on, it is vital that more work is done towards improving our fundamental understanding of the molecular mechanisms of the virus. In this study, the team analyzed the ability of SARS-CoV-2 structural proteins to form VLPs and found that the M protein alone was not sufficient for VLP formation, but co-expression of M with N or S proteins was essential for forming VLPs. They also found that the E protein has an additive effect in the incorporation of N into VLPs as well as in VLP production, which highlights the key role E plays in the assembly and release of the virus.
According to the authors, their findings present a new SARS-CoV-2 viral entry model in a BSL-2 setting: SARS-CoV-2 GFP- and APEX2-VLPs. According to live virus data, GFP-VLPs colocalize with Rab5, the early endosome marker, and LAMP1, the late endosome marker.
“In this work, we also present for the first time a realistic model of SARS-CoV-2 viral entry available in a BSL-2 setting: SARS-CoV-2 GFP- and APEX2-VLPs. In accordance with live virus data, GFP-VLPs colocalize with the early endosome marker, Rab5, and the late endosome marker, LAMP1.”
The team plans to focus their future work on miniaturizing their GFP-VLP entry assay and utilizing it in the screening of viral uptake and entry inhibitors. Apart from using confocal microscopy to evaluate GFP-VLP entry events, they also utilized APEX tagging technology to make it possible to use electron microscopy for the evaluation of SARS-CoV-2 entry.
Traditional approaches to demonstrate VLP-like structures using TEM base their identification solely on morphology. By using APEX tagging, the team was able to show S protein localization during VLP assembly and budding and also the formation and export of APEX-VLPs from the ERGIC lumen.
“In total, this research provides ample resources for other BSL-2 laboratories interested in joining the growing field to try and understand SARS-CoV-2 assembly, budding, and entry dynamics, biochemical and biophysical questions on the four structural proteins, and drug screening of viral assembly, budding, or entry inhibitors.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
SARS-CoV-2 viral budding and entry can be modeled using virus-like particles Caroline B. Plescia, Emily A. David, Dhabaleswar Patra, Ranjan Sengupta, Souad Amiar, Yuan Su, Robert V. Stahelin bioRxiv 2020.09.30.320903; doi: https://doi.org/10.1101/2020.09.30.320903, https://www.biorxiv.org/content/10.1101/2020.09.30.320903v1
- Peer reviewed and published scientific report.
Plescia, Caroline B., Emily A. David, Dhabaleswar Patra, Ranjan Sengupta, Souad Amiar, Yuan Su, and Robert V. Stahelin. 2021. “SARS-CoV-2 Viral Budding and Entry Can Be Modeled Using BSL-2 Level Virus-like Particles.” Journal of Biological Chemistry 296 (January): 100103. https://doi.org/10.1074/jbc.ra120.016148. https://www.jbc.org/article/S0021-9258(20)00093-9/fulltext.