As per the latest WHO reports, coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has infected over 36.37 million people and caused over 1 million deaths across the world.
The virus emerged in Wuhan, China, in late December 2019 and has spread to most countries in a relatively short time. SARS-CoV-2 spreads from person to person through respiratory droplets laden with the virus.
Recent studies show that it is also transmitted through contact on surfaces (fomites), as the virus can remain infectious up to 72 hours - a highly plausible cause for the global spread of the disease. With this information, it is now essential to have a protocol to identify SARS-CoV-2 from surfaces in the built environment efficiently.
Built environments refer to man-made environments that provide the setting for human activity - ranging from buildings, cities, and beyond. These structures include surfaces made of materials such as bare stainless steel, painted stainless steel, plastics, copper, wood, and reinforced fiberglass.
In a recent paper in an open-access journal, mSystems, published by The American Society for Microbiology, Ceth W. Parker et al. show challenges in recovering the virus, change in infectivity, and the case of viral persistence in built environments.
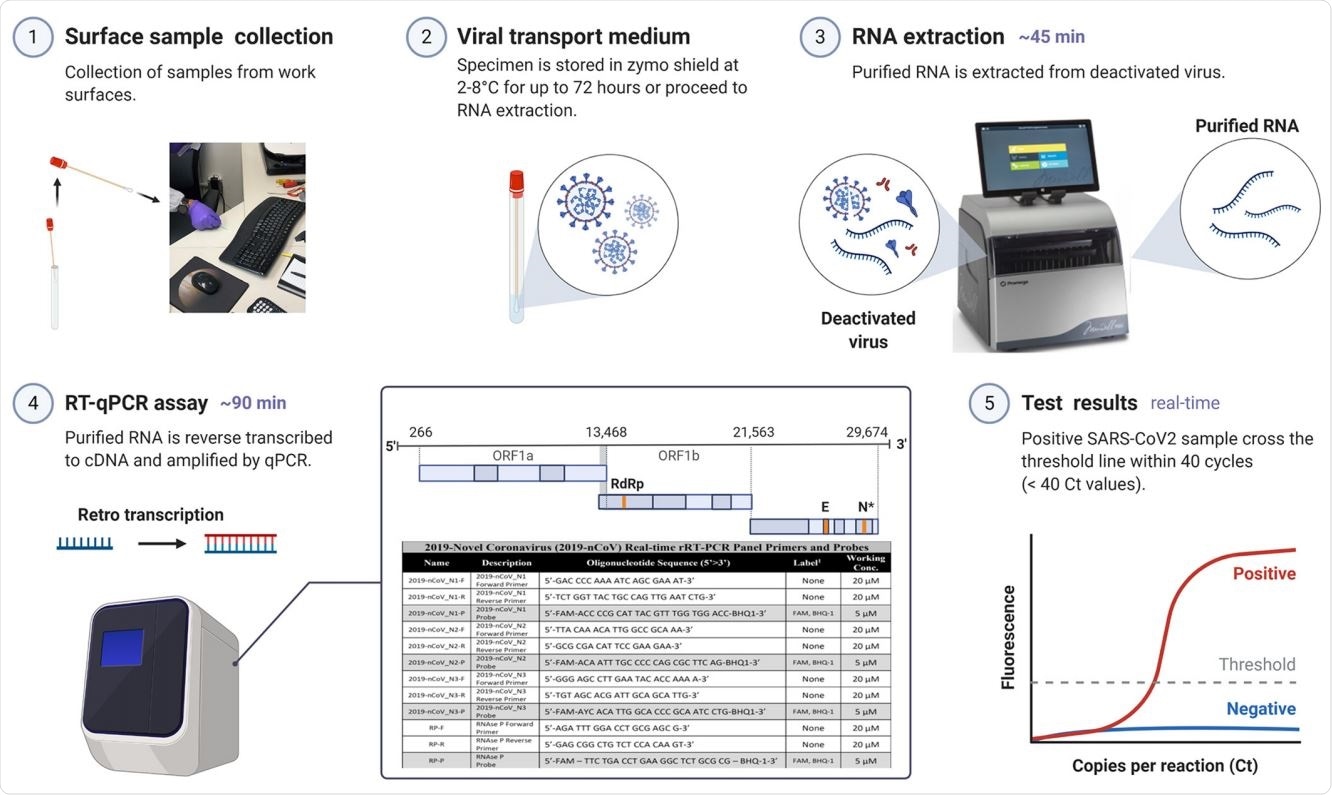
Environmental surface testing using E2E protocol. The optimized E2E protocol for detecting SARS-CoV-2 virus on surfaces is a 5-part procedure: (1) surface sample collection, (2) viral transport medium, (3) RNA extraction, (4) RT-qPCR assay, and (5) test results.
The researchers developed a standardized end-to-end (E2E) protocol for the detection of SARS-CoV-2 from built environmental surfaces and determined the minimum number of RNA copies needed on surface/fomites to positively detect the virus within the limit of detection of the assay used in the study.
"As we all know, SARS CoV-2 is of worldwide concern," said principal investigator of the study Dr. Kasthuri Venkateswaran, a senior research scientist at NASA's Jet Propulsion Laboratory (JPL) in Southern California.
The researchers used a noninfectious and replication-deficient virus used as a surrogate for the SARS-CoV-2 virus. The representative test surfaces inoculated with this virus are: bare 302 stainless steel, painted 302 stainless steel (white acrylic paint), polyethylene terephthalate modified with glycol (PETG), and fiberglass-reinforced plastic (FRP).
To monitor the SARS-CoV-2 on surfaces, the effective combination of methodology included an Isohelix swab collection tool, DNA/RNA Shield as a preservative, an automated system for RNA extraction, and reverse transcriptase quantitative PCR (RT-qPCR) as the detection assay.
"Our group adapted the CDC-approved reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) methodology and then tested the efficacy of RT-qPCR in detecting SARS CoV-2 from various environmental surface samples," said Dr. Ceth W. Parker, a postdoctoral fellow at JPL.
In this comprehensive end-to-end study, the researchers show that the effective combination for monitoring SARS-CoV-2 on surfaces required a minimum of 1,000 viral particles per 25 cm2 for successfully detecting the virus from the surfaces.
When this method was employed to evaluate 368 samples collected from various built environmental surfaces, all the samples turned negative for the viral fragment. It is also found in this study that the debris found on these surfaces negatively impacted the recovery of RNA; Amerstat demonstrated the highest inhibition (>90%) when challenged with an inactivated viral control.
"We tested these surfaces by seeding inactivated noninfectious SARS CoV-2 particles and then determining how well we could actually recover them from the surfaces," said Dr. Parker. "It takes a minimum of 1,000 viral particles per 25 cm2 to effectively and reproducibly detect SARS-CoV-2 virus on the surface. We found that viral RNA can persist on surfaces for at least 8 days. We also found that inhibitory substances and debris have to be taken into account on the surfaces they are being tested on."
Despite the observed longevity of the viral fragment on surfaces, the authors report that the surfaces were either void of virus or below the detection limit of the assay.
The data from this study reflects an overall E2E process efficiency of 0.1%. The E2E assay was also repeated similarly by an independent laboratory for reproducibility and verification purposes.
The results obtained from this evaluation were comparable or equivalent to the results presented in this study.
This detailed study provides necessary protocols for monitoring the SARS-CoV-2 virus on various materials of surfaces and the capacity to retain viral RNA and also allow for effective disinfection.
The information is valuable during this current pandemic - if the virions remain infectious on surfaces for days leading to human infection from contact with infected surfaces?
Because no policy is set for environmental surface monitoring of the virus, a thorough approach is outlined in this study to characterize and develop a practical methodology to understand the viral persistence in built environments.
Such intensive research on SARS-CoV-2 is essential to understand the pathogenic mechanisms and epidemiological behavior - these will help develop effective preventive and therapeutic strategies.
Journal reference:
- End-to-End Protocol for the Detection of SARS-CoV-2 from Built Environments Ceth W. Parker, Nitin Singh, Scott Tighe, Adriana Blachowicz, Jason M. Wood, Arman Seuylemezian, Parag Vaishampayan, Camilla Urbaniak, Ryan Hendrickson, Pheobe Laaguiby, Kevin Clark, Brian G. Clement, Niamh B. O'Hara, Mara Couto-Rodriguez, Daniela Bezdan, Christopher E. Mason, Kasthuri Venkateswaran mSystems Oct 2020, 5 (5) e00771-20; DOI: 10.1128/mSystems.00771-20, https://msystems.asm.org/content/5/5/e00771-20