An international team of scientists has developed computational models to re-analyze and validate experimentally-derived publicly available macromolecular structures of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). They have created a website to deposit structural models that they have improved by automatic and manual evaluation. The study is currently available on the bioRxiv* preprint server.
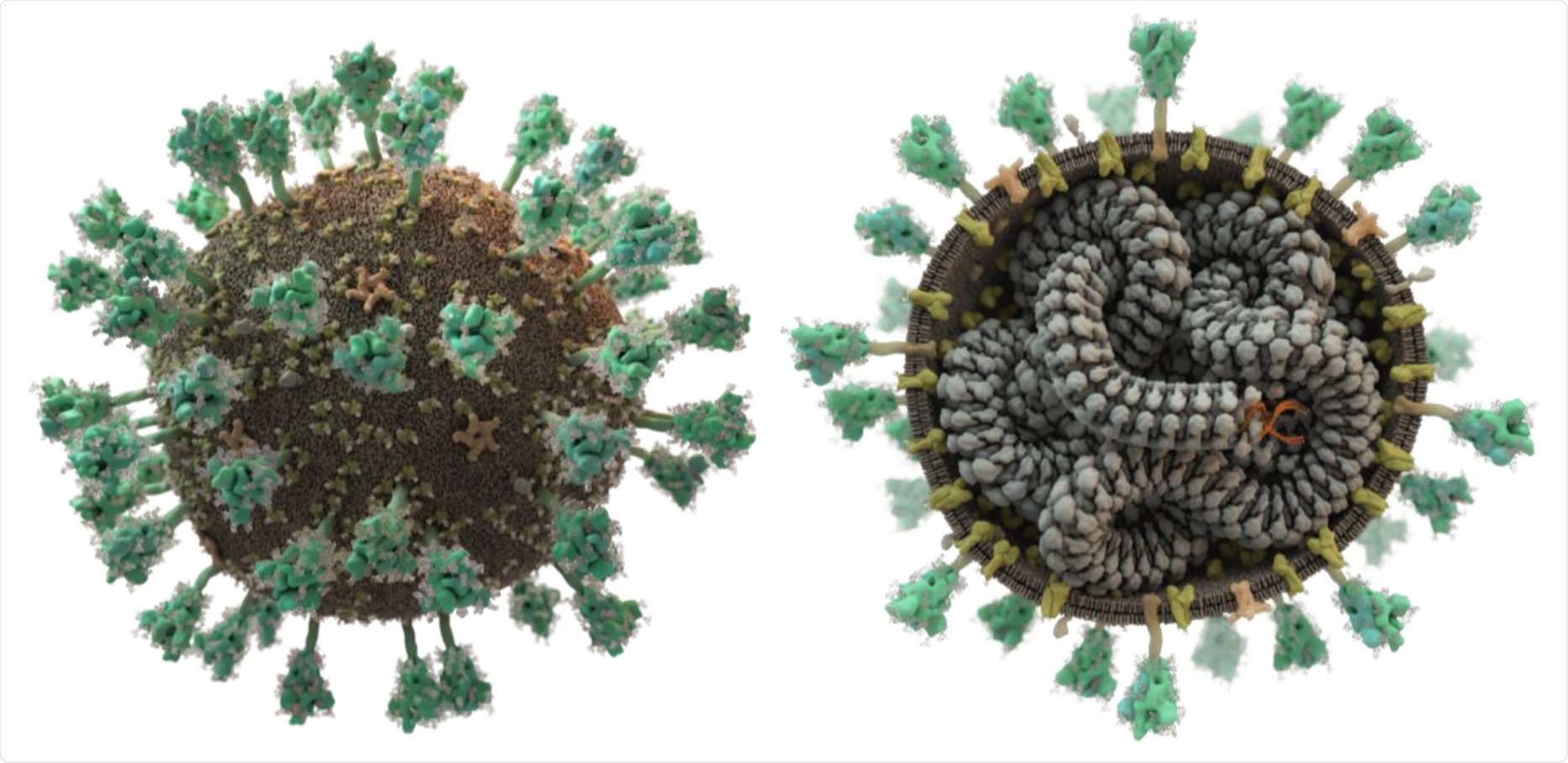
SARS-CoV-2 displays spike proteins (green) on its surface that recognise and bind to host cells; its lipid bilayer membrane also contains additional embedded membrane and envelope proteins (yellow and beige). The single-stranded RNA (orange) is intertwined in a helical fashion with the nucleocapsid (grey). This figure, however, shows only the transport form of the virus: once a cell is infected, additional viral proteins encoded by the viral RNA are produced that hijack the host cell in order to produce new virus particles. (Picture: Thomas Splettstößer /scistyle.com)

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
SARS-CoV-2, the causative pathogen of coronavirus disease 2019 (COVID-19), is a positive-sense, single-stranded RNA virus with a genome size of 30 kb. The SARS-CoV-2 genome encodes a total of 28 proteins that are essential for viral transmissibility, replication, survival, and host immune evasion. Therefore, structural and functional characterization of these proteins is of prime importance to thoroughly understand the viral life cycle and identify potential therapeutic targets.
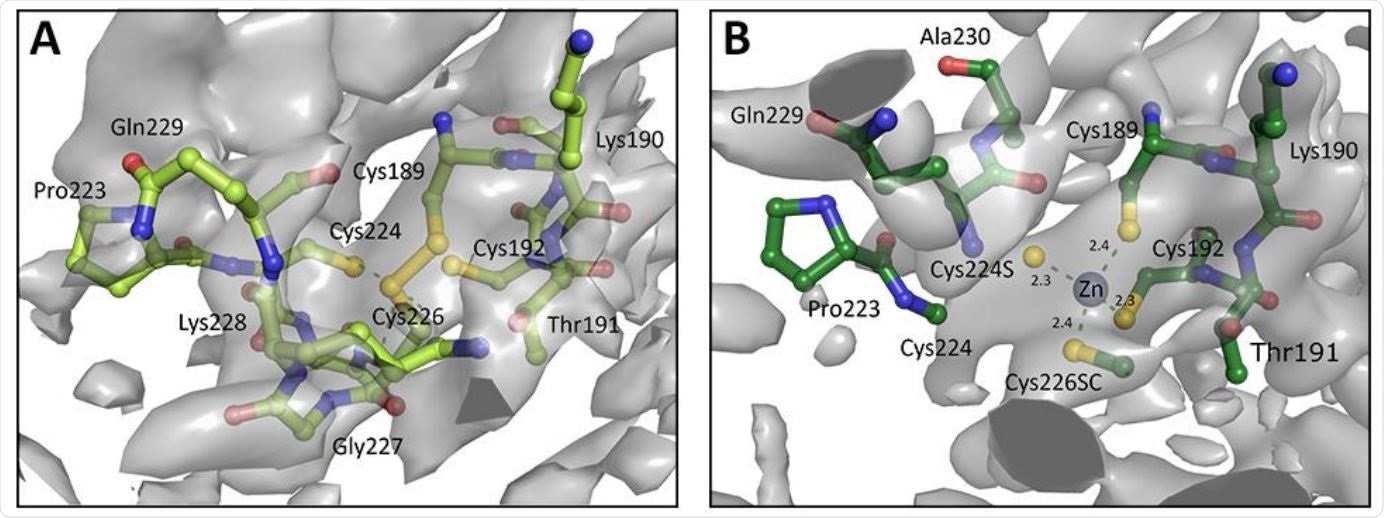
A. Chain A of zinc finger from PDB entry 6W9C as deposited, with Cys189 and Cys226 forming a disulphide bond instead of a Zn binding site. B. Re-modelled structure with zinc binding site, utilising 3-fold NCS, prior knowledge about coordination chemistry, and increased geometry weights to improve the map. Electron density is displayed as an isosurface contoured to 1σ.
Since the emergence of the COVID-19 pandemic, a considerable number of studies have been undertaken to develop atomic structures of these viral proteins using nuclear magnetic resonance, cryo-electron microscopy, and crystallographic techniques. Scientists make these structures freely and publicly available in the World Wide Protein Data Bank (wwPDB) to benefit up-coming research related to the COVID-19 pandemic. Within the last 6 months, a total of 367 macromolecular structures covering 16 proteins of SARS-CoV-2 have been deposited. Because of the immense pressure of fast-paced research, mistakes frequently occur even in very carefully derived macromolecular structures. As these structures are used to evaluate important viral functions, even a small mistake can cause serious consequences. Therefore, accurate validation of publicly available structures is an absolute requirement for successfully combating SARS-CoV-2.
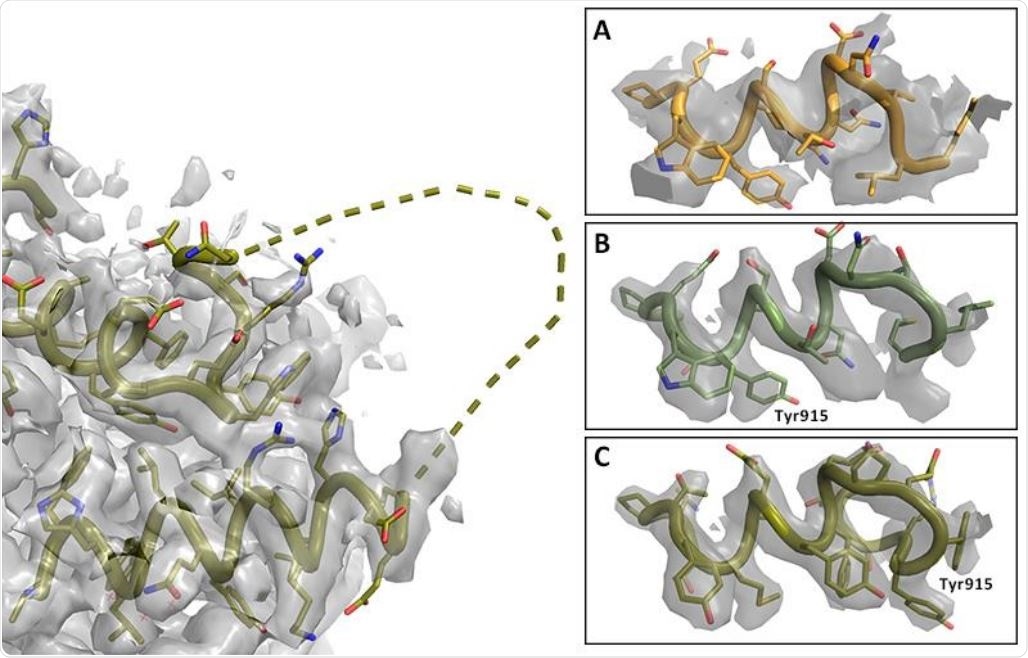
Registry shift in C-terminus of RNA Polymerase. Left: Overview with missing loop shown as dashed line (PDB entry 7BV2); map at 2.4σ. Right: Details of C-terminal helix at 5σ. A. Lower resolution map and model PDB 6NUS. Judging the side chain fit is difficult. B. Higher resolution map and model 7BV2 as deposited; the side chain fit is suboptimal. C. Amended 7BV2 structure; the side chains now fit the density. The register shift is indicated by Tyr915.
Current study design
The scientists developed computational methods to re-analyze and validate publicly available macromolecular structures of SARS-CoV-2. All representative structures underwent automatic post-analysis and manual re-processing and re-modeling. The website (insidecorona.net) they created contains significantly improved macromolecular models of many SARS-CoV-2 proteins, which are made freely and publicly available and have been used widely by scientific communities.
In automatic validation, SARS-CoV and SARS-CoV-2 related macromolecular structures are downloaded into the repository and analyzed automatically within 24 hours of release. For cryo-electron microscopic and crystallographic structures, the quality of deposited merged data is checked initially, followed by re-analysis of structures based on prior chemical knowledge.
For crystallographic data, they checked for twinning, completeness, and overall diffraction quality using computational models. They observed that 7 of 415 datasets have completeness below 80%. About 61 datasets showed ice rings, and 49 crystal structures were found to have resulted from twinned crystals.
They also checked how atomic models fit the data. They observed significantly high R-value (a quality measure) of more than 35% for two structures, which they improved using PDB-REDO, a procedure to optimize crystallographic models.
After analyzing cryo-electron microscopic structures, they observed that 6 of 81 deposited structures had bad overall fit between the model and map. For 12 structures, a poor fit of more than 5% of residues with the map was observed.
To validate the structures based on prior chemical knowledge, they checked for covalent geometry, conformational parameters of protein and RNA, and steric clashes. They observed that for many structures, the backbone conformations were incorrect.
For manual evaluation, they selected representative structures and found that the most common errors were peptide bond flips, rotamer outliers, and misidentification of small molecules. Of all manually checked structures, 31 were improved significantly and made freely available on the website.
Study significance
According to the scientists, the major problem with wwPDB is the unavailability of raw data, which are essential for re-analysis and validation of existing models and the development of new models. To maximize the utility of experimental results, the current study scientists invited other scientists to deposit their raw data so that an analytical platform can be created for re-analysis and validation of viral structural models.
By continually validating and updating viral structures, the scientists aim at constantly improving the outcomes of new research.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Tristan Croll, Kay Diederichs, Florens Fischer, Cameron Fyfe, Yunyun Gao, Sam Horrell, Agnel Praveen Joseph, Luise Kandler, Oliver Kippes, Ferdinand Kirsten, Konstantin Müller, Kristopher Nolte, Alexander Payne, Matthew G. Reeves, Jane Richardson, Gianluca Santoni, Sabrina Stäb, Dale Tronrud, Christopher Williams, Andrea Thorn, bioRxiv 2020.10.07.307546; doi: https://doi.org/10.1101/2020.10.07.307546, https://www.biorxiv.org/content/10.1101/2020.10.07.307546v1
- Peer reviewed and published scientific report.
Croll, Tristan I., Kay Diederichs, Florens Fischer, Cameron D. Fyfe, Yunyun Gao, Sam Horrell, Agnel Praveen Joseph, et al. 2021. “Making the Invisible Enemy Visible.” Nature Structural & Molecular Biology 28 (5): 404–8. https://doi.org/10.1038/s41594-021-00593-7. https://www.nature.com/articles/s41594-021-00593-7.