An international team of scientists has identified novel T cell-recognized epitopes spanning different proteins of severe acute respiratory syndrome coronavirus 2 in COVID-19 recovered patients. The scientists have also revealed phenotypical characteristics of the T cell population. The study is currently available on the bioRxiv* preprint server.
Since its emergence in December 2019, deadly severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative pathogen of coronavirus disease 2019 (COVID-19) pandemic, has infected more than 37.3 million people and claimed more than one million lives worldwide. A wide variety of clinical consequences have been observed in COVID-19 patients, ranging from asymptomatic or mild infection to severe or life-threatening infection.
A diverse range of immune responses to SARS-CoV-2 have been observed, wherein higher antibody titers are associated with male gender, age, and hospitalization status. Many studies have shown that T cell-mediated immune response against SARS-CoV-2 may provide long-term protection. In COVID-19 recovered patients, a robust SARS-CoV-2-specific T cell response has been observed. Similarly, a robust CD8+ T cell-mediated immune response has been observed in COVID-19 patients with mild symptoms.
Given the significant involvement of T cells in mitigating disease severity, it is essential to study the specific targets of T cells in the viral proteome and the clinical consequences of T cell-mediated targeting.
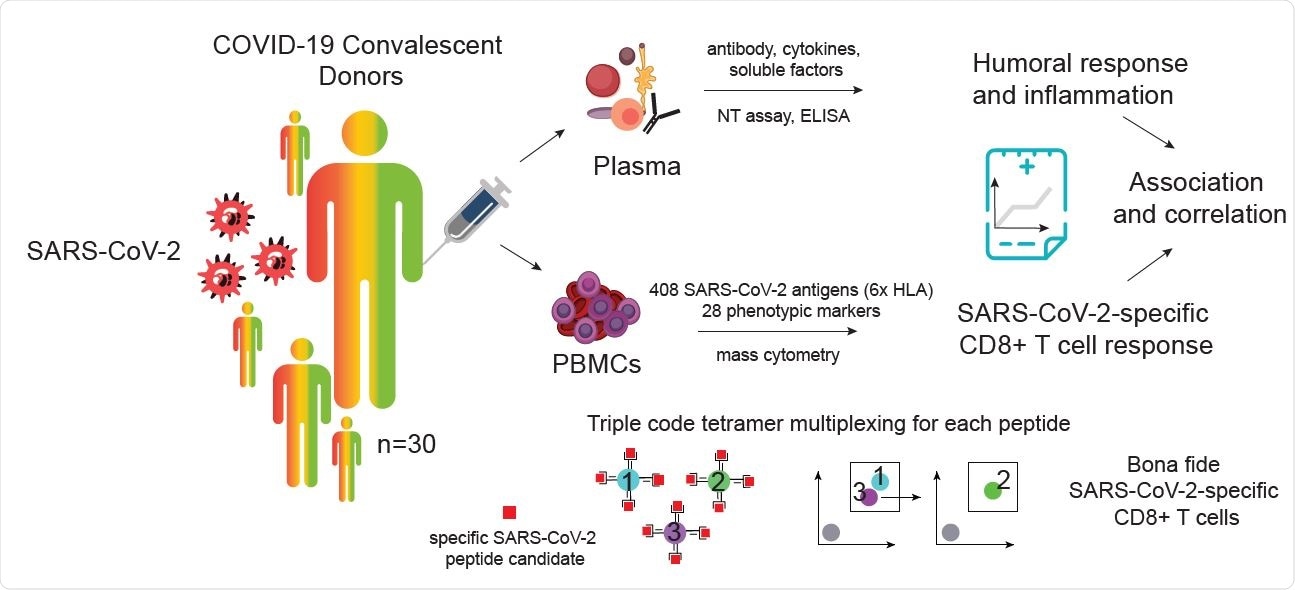
Identification and characterization of SARS-CoV-2-specific CD8+ T cells from SARS-CoV-2 convalescent donors. A) Visualization and schematic overview of the experimental workflow. SARS-CoV2-specific CD8+ T cells were identified and simultaneously characterized in PBMCs from convalescent donors by screening a total of 408 SARS-CoV-2 candidate epitopes across six HLAs using a mass cytometry based highly multiplexed tetramer staining approach. Frequencies and phenotypic profiles of SARS-CoV-2-specific T cells were associated and correlated with the cross-sectional sample-specific humoral response and inflammation parameters.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Current study objective
In the current study, the scientists aimed to identify CD8+ T cell-recognized epitopes in COVID-19 recovered patients spanning different proteins of SARS-CoV-2. They also evaluated the phenotypic characteristics of these CD8+ T cells.
Important observations
The scientists analyzed 30 convalescent plasma samples (donor age range: 19 – 77 years) to identify and characterize the SARS-CoV-2-specific CD8+ T cell population using a mass cytometry-based multiplexed tetramer staining approach. To associate T cell response with antibody response, they analyzed several plasma-derived parameters, such as IgG level, neutralizing antibody level, and inflammatory mediator level. The results showed that the anti-spike protein IgG level correlates strongly with neutralizing antibody activity.
They screened 408 potential epitopes for CD8+ T cells across 6 human leukocyte antigen (HLA) alleles. This led to the detection of 52 unique hits (peptide reactivity) distributed across 132 epitopes of SARS-CoV-2. Almost all donors showed CD8+ T cell-mediated immune responses, and more than 40% of donors showed more than 5 different SARS-CoV-2 specificities. Also, they observed that most of the unique hits were directed against not-structural SARS-CoV-2 proteins, such as NSP, PLP, and ORF3a.
The most common hits were detected against the spike protein and ORF3a; however, the frequency of nucleocapsid-specific CD8+ T cells was significantly higher than that of spike- and non-structural protein-specific T cells. Overall, 6 out of 14 different epitopes (per HLA allele) showed T cell-specific reactivity. These findings indicate that CD8+ T cells of COVID-19 recovered patients can recognize a broad range of epitopes spanning different viral proteins.
Regarding phenotypic characterization of T cells, the scientists observed that the majority of SARS-CoV-2-specific T cells share similar profiles, which were different from profiles of influenza-, Epstein Barr Virus-, and cytomegalovirus-specific T cells (control group). Moreover, SARS-CoV-2-specific T cells showed stem-cell memory and transitional memory cell-2 (TM2) phenotypes.
Regarding T cell expansion, high prevalence SARS-CoV-2-specific T cells were associated with late differentiation, whereas low-prevalence SARS-CoV-2-specific T cells were associated with early differentiation. T stem-cell memory cells' capability to differentiate into various subsets of T memory cells may be associated with long-term protection against SARS-CoV-2 in COVID-19 recovered patients.
Regarding the relationship between inflammation, antibody-mediated immune response, and T cell-mediated immune response, the scientists observed an increased SARS-CoV-2-specific T cell response during the resolution phase of COVID-19. Moreover, higher frequencies of T cells with terminal differentiation and activation markers were correlated with longer recovery time. This indicates that the proliferation and progressive differentiation of SARS-CoV-2-specific T cells correlate with recovery time. A negative correlation between inflammation and recovery time was indicated by a reduction in plasma cytokine levels with time.
Overall, the study provides new insights into SARS-CoV-2-specific T cell response patterns in COVID-19 recovered patients, which can help identify novel viral targets for drug discovery or vaccine development.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources