Researchers in the United States have developed a computational method called “weighted-nearest neighbor” (WNN) analysis that can significantly improve the ability to define cellular states across various biological contexts and data types.
By applying this analytical framework to a dataset of hundreds of thousands of human white blood cells and more than 200 antibodies, the team created a multimodal atlas of the circulating immune system.
This enabled the researchers to identify heterogeneous cell states in human lymphocytes and investigate immune responses to vaccination and infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the agent responsible for the current coronavirus disease 2019 (COVID-19) pandemic.
A pre-print version of the paper is available on the server bioRxiv*, while the article undergoes peer review.
Limitations of using transcriptomics alone
While established technologies such as single-cell RNA-seq (scRNA-seq) enable the discovery of new cell types and states in heterogeneous tissues, it is often not possible to separate molecularly similar, but functionally distinct, types of immune cells using transcriptomics alone.
Multimodal analysis, which enables the different cellular modalities to be measured simultaneously, could provide a solution to overcoming the limitations of single-cell genomics and help to investigate how multiple modalities affect cellular state and function.
However, such approaches require new computational methods capable of defining cellular states based on multiple different data types.
“For example, while CITEseq [Cellular Indexing of Transcriptomes and Epitopes by Sequencing] datasets can be analyzed by first identifying clusters based on gene expression values, and subsequently exploring their immunophenotypes, a multimodal computational workflow would define cell states based on both modalities,” say Satija and team.
However, it is essential that such strategies are robust to present significant differences in the data quality and content of individual modalities. Variations in the information content between modalities present a challenge in analyzing and integrating multimodal datasets.
What did the researchers do?
Now Satija and colleagues have introduced WNN analysis, which they developed to learn about the relative utility of the data types in each cell and enable an integrative analysis of multiple modalities.
“By calculating cell-specific modality weights, WNN analysis solves an important technical challenge for the analysis of multimodal datasets and allows for flexible application across a range of modalities and data types,” they explain.
The researchers applied this method to a CITE-seq dataset of 211,000 human peripheral blood mononuclear cells (PBMCs) alongside a panel of 228 antibodies to generate a multimodal reference atlas of the circulating immune system.
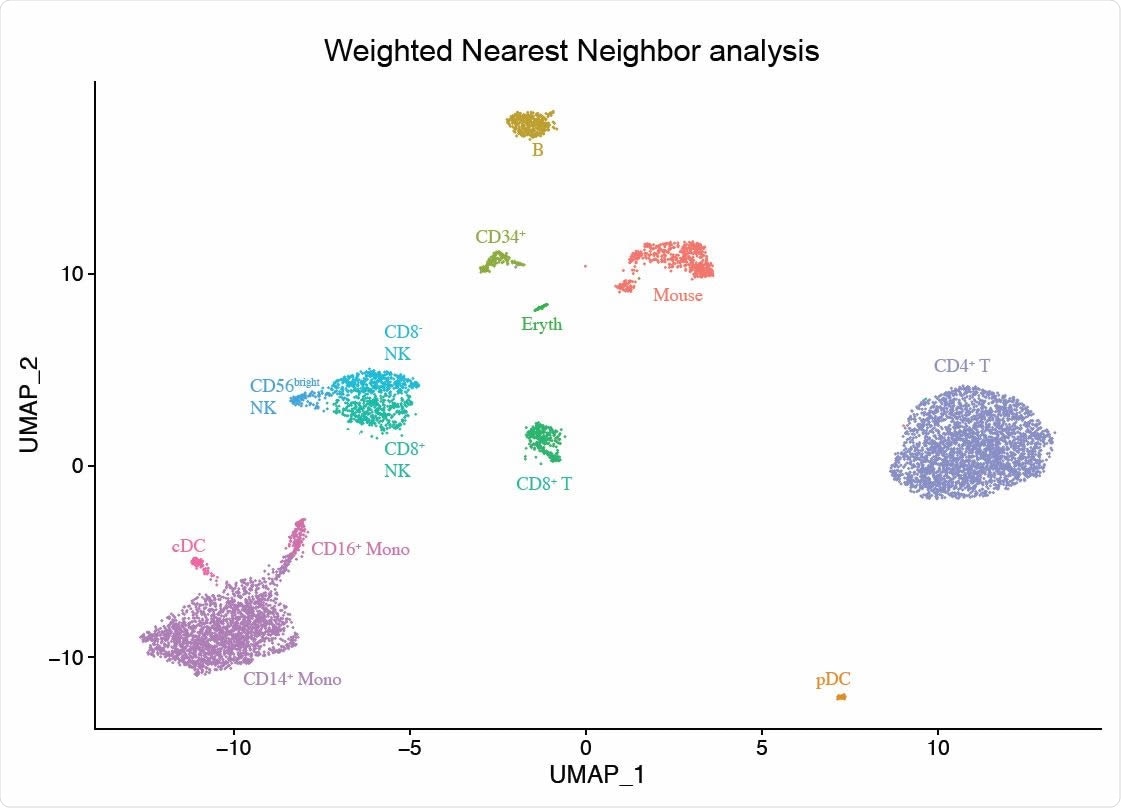
Integrated modalities by constructing a Weighted Nearest Neighbor (WNN) graph, based on a weighted average of protein and RNA similarities. UMAP visualization and clustering of this graph.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
What did the study find?
The team showed that the WNN analysis significantly improved the ability to resolve cellular states in multiple biological contexts and data types and validated the presence of previously unreported subpopulations of lymphocytes.
The researchers observed extensive lymphoid heterogeneity that has not yet been seen using scRNA-seq alone, including differential expression of integrins on circulating memory T cells and tightly clustered clonal populations within groups of effector cells and cytotoxic cells.
Furthermore, this reference atlas enabled them to explore how the innate immune system responds to vaccination, highlighting specific response biomarkers and heterogeneous responses of dendritic cells.
“Importantly, we demonstrate that CITE-seq data can be easily mined to identify the best immunophenotypic marker panels for any subpopulation of interest,” say Satija and colleagues. “These marker panels can be used for flow cytometry with the same antibody clones in our CITE-seq panel, facilitating rapid enrichment and downstream analysis of these groups, and broadening the value of our resource.”
The technique also enabled the researchers to investigate how the innate immune system responds to infection with SARS-CoV-2.
On applying the mapping approach to a recent scRNA-seq study of PBMC samples taken from hospitalized COVID-19 patients, the researchers observed a reduction in the level of mucosal-associated invariant T cells (MAIT cells) among the COVID-19 samples, compared with healthy controls.
“This change in abundance may reflect these cells exiting circulation to play protective roles in barrier tissues during the antiviral immune response,” suggests the team.
Moving beyond the transcriptome towards a multimodal definition of cellular identity
Satija and colleagues say the approach represents a broadly applicable strategy for analyzing single-cell multimodal datasets to move beyond the partial and transcriptome-focused view of a cell towards an integrative, multimodal definition of cellular identity, behavior, and function.
To assist the community in using this resource, the team has created a web application, freely accessible a:t http://www.satijalab.org/azimuth.
“Using this approach, a dataset of 50,000 cells can be fully processed and mapped in less than five minutes,” say the researchers.
“As the profiling of human PBMC under a variety of disease states becomes increasingly routine, the ability to perform automated mapping of these datasets will facilitate the characterization of complex immune responses, and the discovery of pathogenic populations,” they conclude.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Satija R, et al. Integrated analysis of multimodal single-cell data. bioRxiv, 2020. doi: https://doi.org/10.1101/2020.10.12.335331
- Peer reviewed and published scientific report.
Hao, Y., Hao, S., Andersen-Nissen, E., Mauck, W. M., Zheng, S., Butler, A., Lee, M. J., Wilk, A. J., Darby, C., Zager, M., Hoffman, P., Stoeckius, M., Papalexi, E., Mimitou, E. P., Jain, J., Srivastava, A., Stuart, T., Fleming, L. M., Yeung, B., & Rogers, A. J. (2021). Integrated analysis of multimodal single-cell data. Cell, 184(13). https://doi.org/10.1016/j.cell.2021.04.048. https://www.cell.com/cell/fulltext/S0092-8674(21)00583-3.