With the resurgence of the COVID-19 pandemic looming large in many parts of the world, the need for immunological tools such as monoclonal antibodies (mAbs) is more urgent, whether for diagnosis, treatment, or research on the virus. Now, a recent study published on the preprint server bioRxiv* in October 2020 reports the development of a rapid process for generating high-affinity mouse mAbs, and their functional attributes.
The focus of most antibody development efforts is the receptor-binding domain (RBD), a region in the S1 subunit of the SARS-CoV-2 spike protein, about 300 residues in length. This spike protein is thought to be the viral element that mediates its attachment to the host cell through the human angiotensin-converting enzyme 2 (hACE2) receptor. This is found on a variety of cell types in the human host, particularly the lung epithelium.
The RBD contains five residues that come into contact with the ACE2 receptor. In order to exploit this viral element, the researchers used a commercially available S1 domain tagged with a C-terminal histidine (His-S1), a commercial RBD with a mouse C-terminal IgG1 Fc domain (mFc-RBD), and mFc-RBD fused with the PP7 bacteriophage virus-like particle (VLP), called PP7zz.
The latter is modified such that two sequential Z-domains are displayed with each binding to Fc at 120 places on the surface of the VLP. It has a large display of RBDs that enable binding multiple receptors on immune cells, thus allowing the cell affinity to mature.
Any antigen with an Fc tag promotes enhanced antigen uptake and processing. The VLPs have an adjuvant effect, but another adjuvant also enhanced the recombinant S1 and RBD proteins' immunogenicity.
Accelerated Immunization and mAb Production
The researchers used accelerated immunization schedules using either the His-S1, a combination of mFc-RBD and PP7zz immunogens, and mFc-RBD expressed on VLPs boosted finally by His-S1. The aim was to find which immunogen displayed the maximum response. Spleen cells were then harvested on day 30 from immunization, and cells that secreted high IgG levels were chosen.
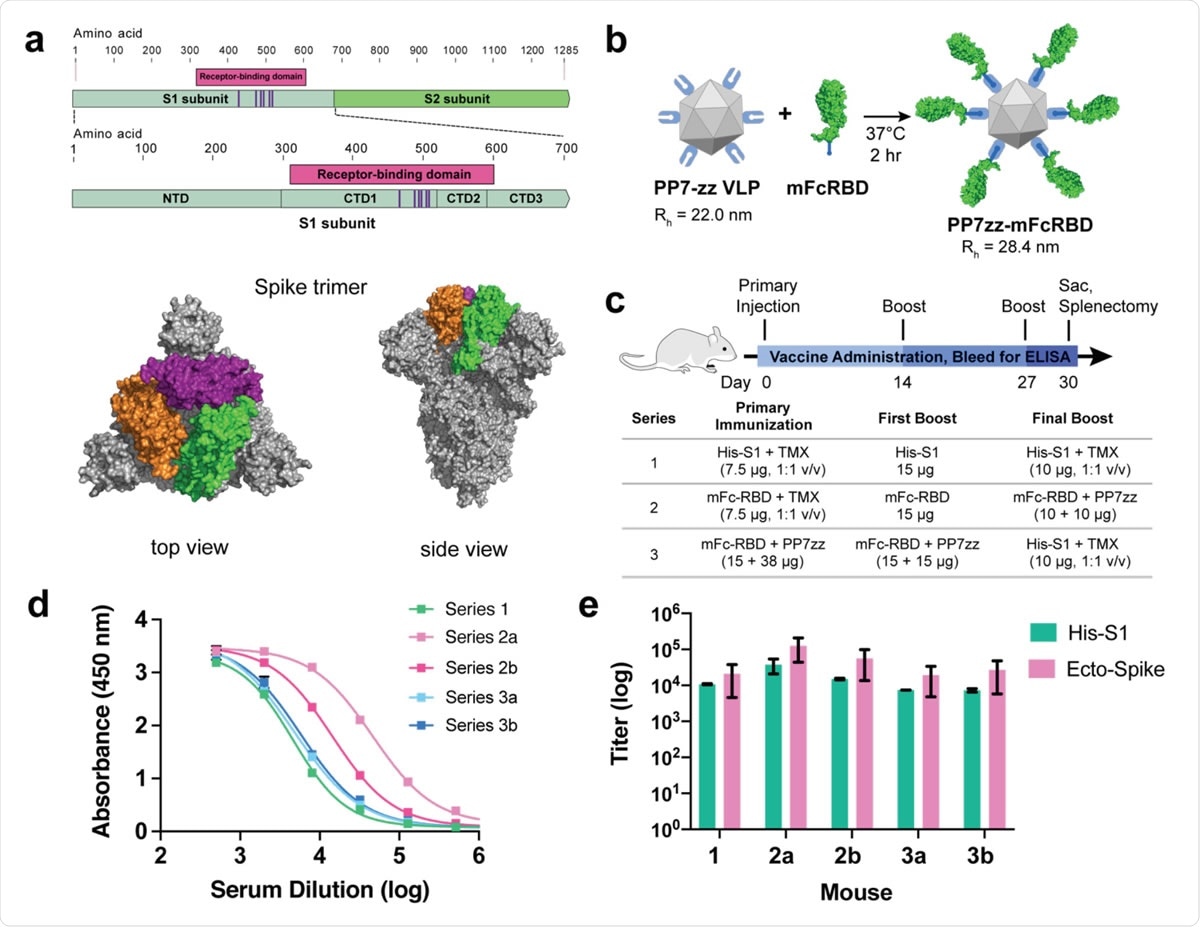
SARS-CoV-2 Spike protein subunit vaccine strategy and humoral immune response in mice. a, Recombinant spike subunit 1 protein (His-S1, residues 1-681) or S1 Receptor Binding Domain (mouse Fc-RBD, residues 319-541, ACE2 contact residues in purple; PDBID: 6vxx) antigens. b, VLP display of Fc-tagged antigens using the PP7 particle bearing 120 ZZ-domains; a 1:1 mass ratio of mFc-RBD and VLP provides a Fc:ZZ molar ratio of approximately 0.8. Rh = hydrodynamic radius measured by dynamic light scattering in phosphate buffer. c, Vaccine schedule and strategy. Six-week old female BALB/c mice (n = 3 per group) were immunized with primary antigen and adjuvant on day 0 followed by boosts on days 14 and 27. Blood was collected for ELISA on days 0, 14, 21 and 30. d, ELISA responses for serum dilutions against plated His-S1 protein from the sacrificed mice at day 30. e, Titer values from ELISA analysis as in panel d, against plated His-S1 or spike ectodomain protein. Immunization series defined in panel c; a and b designate different mice within that series. Experimental error represents standard deviation.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Characterization of Antibodies by Function
The antibody titer was then assayed by ELISA, using either recombinant His-S1 or engineered SARS-CoV-2 spike protein, the first being a monomer and the second a trimer.
This showed the high immunogenicity of the mFc-RBD antigen, which generated spike-binding mAbs. These were sorted, and the selected mAbs were characterized for binding strength, the avidity of binding, and affinity.
The researchers found 33 selected avidly binding light chain IgG1 molecules, irrespective of the spike target protein's storage or incubation under various conditions.
The mAbs were then clustered according to the differential expression of various gene sets. The researchers found that neutralizing capacity differentiated the mAbs into three groups, 1-3, which were related to the presence of IGHV1 and 8, and IGKV1, 3, and 6. Group 3 is a large group of non-neutralizing antibodies, all of which are associated with the IGHV14-1 gene and, in 50% of mAbs, IGKV6-32.
Evaluating Epitopes Recognized by the mAbs
Competitive binding measurements are important in evaluating the epitopes involved in spike binding by the antibodies. Low competitive binding values showed the antibodies were competing for adjacent or the same residues, or residues which were allosterically related.
On the other hand, high values showed antibody association to different epitopes. This assay showed six major binding groups related to heavy chain sequences and specific combinations of IGHV and IGKV genes.
Impact on RBD-ACE2 Binding
They also measured the effect of these mAbs on RBD-ACE2 binding, both directly and indirectly. They found 10 mAbs, three of which had identical sequences, which showed strong RBD binding inhibition with both the full-length ACE2 and a truncated receptor version that does not present the dimerization domain of the ACE2 molecule.
They observed potent neutralization with 8 of these 10 antibodies, as well as another two with a different mechanism of inhibition of RBD-ACE2 binding. However, of the mAbs, 21 caused antibody-dependent enhancement (ADE) with the truncated ACE2 but not the full-length ACE2, in 17/21 antibodies. These, therefore, have different recognition epitopes and belong to a different functional class than the first 10.
Binding to Denatured RBD
Using a luciferase assay, they found some variations in binding patterns, which indicate that these ten antibodies bind to four or more RBD epitopes. Four of them bind specifically to a small peptide on the RBD that contains four of five contact residues. However, none of these were found to have potent neutralizing ability, which may mean that this peptide in linear form does not resemble that present in the infectious viral particle, but only on degraded virions, as shown by the binding of two of these mAbs to spike protein in lung tissue sections.
The same pattern was seen following gamma-irradiation of the virions, where mAbs with similar avidity of binding for the spike protein showed comparable neutralizing power in vitro. Still, one had a limit of detection of inactivated virions, which was an order of magnitude larger than the other.
Implications
This study shows that the use of Fc- and VLP-assisted RBD antigens for immunization combined with rapid hybridoma production of the mAbs in parallel can present a range of binding antibodies that have different but useful functional characteristics for binding and inhibiting the virus. This agrees and fills out earlier studies where B cells from COVID-19 patients were used to produce a large array of mAbs targeting SARS-CoV-2.
As in the prior studies, the current approach successfully elicited a number of mAbs with affinities in the low nanomolar range for the spike protein. This confirms many earlier observations that the RBD is a useful target for immunological therapeutic agents.
Overall, the study shows at least five functional groups of antibodies. Three groups comprise neutralizing antibodies that recognize distinct epitopes on the RBD. A fourth represents the large group of non-neutralizing mAbs, and the fifth includes mAbs that bind to denatured or inactivated RBD in infected lung tissue samples. Each functional class seems to have distinct neutralizing potency, different epitope recognition sites, and sequence and varying RBD-ACE2 binding effects.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources