Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of coronavirus disease 2019 (COVID-19), and it primarily infects cells in the respiratory tract. The human airway epithelium is the first site of infection by SARS-CoV-2. The cellular tropism of the SARS-CoV-2 virus affects several aspects of infection, such as the spread of the virus within and between hosts, tissue pathology, immune control mechanisms, and the response to treatment with promising antiviral drugs.
Human tracheal bronchial epithelial cells represent a heterogeneous mix of ciliated cells, basal cells, and secretory cells. When cultured at the air-liquid interface, these cells form a pseudostratified epithelium, mimicking the upper airway in humans. Interestingly, cells in this culture also express endogenous levels of crucial host factors such as angiotensin-converting enzyme 2 (ACE2) and proteases like TMPRSS2, which are essential for the entry of SARS-CoV-2 into the host cell membrane.
Several recent studies used primary lung cell cultures and respiratory cells from patients with SARS-CoV-2 infection and demonstrated SARS-CoV-2 viral tropism for secretory and ciliated cells in the upper airway. However, these studies failed to throw light on the heterogeneity of SARS-CoV-2 replication and induction of antiviral genes and proinflammatory cytokines in these cells.
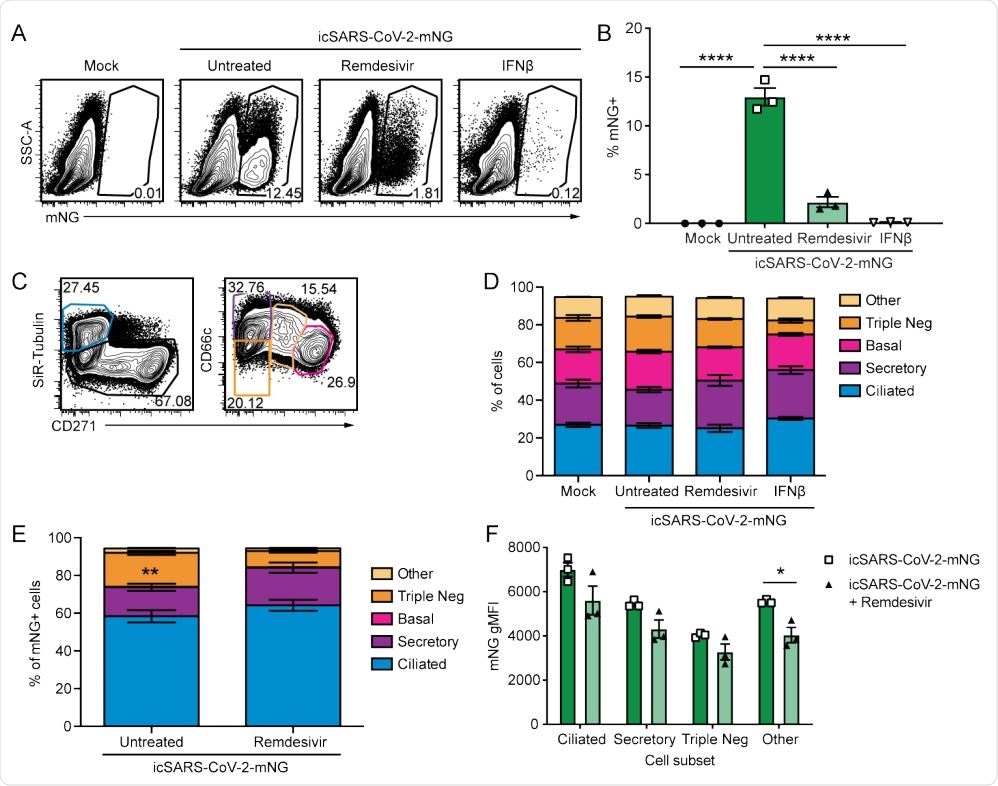
. SARS-CoV-2 infects multiple cell types and infection is controlled by remdesivir in primary nHTBE cells. nHTBE cells differentiated at air-liquid interface were untreated or treated with IFN-β or remdesivir prior to infection with icSARS-CoV-2-mNG at an MOI of 2.5. Cells were analyzed 48 hpi by flow cytometry. (A) Representative plots of live cells (B) Frequency of infected cells (mNG+ ). (C) Representative plots of cell subset gating. Ciliated (SiR-Tubulin+ CD271- ), secretory (SiR-Tubulin- CD271- CD66c+ ), basal (SiR-TubulinCD271+ CD66c- ), Triple-negative (SiR-Tubulin- CD271- CD66c- ) and other cells (SiR-Tubulin- CD217int CD66cint) (D) Frequency of cell subsets from live cells. (E) Frequency of cell subsets of infected cells (mNG+ ). (F) mNG gMFI within each cell subset. The data (B, D-F) are 3 biological replicates per group +/-SEM, 1 of 2 independent experiments.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Single-cell RNA sequencing reveals SARS-CoV-2 tropism and induction of antiviral immune responses
Researchers from the University of Minnesota and the University of Texas Medical Branch recently defined SARS-CoV-2 tropism and the induction of antiviral and pro-inflammatory immune responses using single-cell RNA sequencing. Their study is published on the preprint server bioRxiv*.
The researchers also used flow cytometry to analyze how the airway's diverse cell population contributes to viral tropism and pathogenesis, antiviral immune response, and treatment response to the drug, remdesivir.
"Here, we use nHTBE cells infected with SARS-CoV-2 to demonstrate that remdesivir reduces viral replication uniformly in all susceptible cell types within the upper respiratory tract."
SARS-CoV-2 predominantly targets ciliated airway epithelial cells
The researchers found that, though many cell types in the airway epithelium are susceptible to SARS-CoV-2 infection, ciliated cells are prime targets for the virus.
In this study, treatment with remdesivir successfully inhibited replication of SARS-CoV-2 across many cell types, and remdesivir also dampened hyperinflammatory responses. The team also found that highly infected epithelial cells show impaired IFN signaling. These cells expressed plenty of IL-6, which is a mediator of COVID-19 pathogenesis.
"Together, the results presented here demonstrate that ciliated airway epithelial cells are the predominant cell type initially infected by SARS-CoV-2 and reveal heterogeneity within infected cells and in antiviral responses."
Broad tropism and poor virus sensing likely allowed zoonotic emergence of SARS-CoV-2
As the COVID-19 pandemic continues to spread across the globe, causing significant morbidity and mortality, understanding the virus-host interaction's initial steps in relevant models is key to grasping the disease's pathophysiology. It will also help evaluate antiviral and immunomodulatory therapeutics.
In this study, the research team demonstrated that the SARS-CoV-2 virus prefers ciliated and secretory cells in the upper airway of humans. However, among these primary cells, the virus is only sensed by a few infected cells leading to activation of IFN. Despite this, a potent antiviral immune response is established.
Interestingly, the authors discovered that at 48 hours post-infection, TMPRSS2 expression, and IFN responses are the major determinants of SARS-CoV-2 tropism in the susceptible cells. According to the team, their results show that infected epithelial cells can produce IL-6 at the early stages of infection, which likely contributes to the inflammatory cascade that follows at later stages of infection.
The study also found that remdesivir can inhibit the replication of the virus across all susceptible cell types in the upper airway. Zooming in on the virus-host interactions at the single-cell level shows that broad tropism of SARS-CoV-2 and successful evasion of virus sensing by the cells may be the key factors that might have facilitated the zoonotic emergence of the virus.
"Defining virus-host interactions at the single-cell level reveals broad tropism and successful evasion of virus sensing, key attributes that likely allowed the zoonotic emergence of SARS-CoV-2."

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Single-cell resolution of SARS-CoV-2 tropism, antiviral responses, and susceptibility to therapies in primary human airway epithelium Jessica K. Fiege, Joshua M. Thiede, Hezkiel Nanda, William E Matchett, Patrick J. Moore, Noe Rico Montanari, Beth K Thielen, Jerry Daniel, Emma Stanley, Ryan C Hunter, Vineet D Menachery, Steven S. Shen, Tyler D. Bold, Ryan A. Langlois bioRxiv 2020.10.19.343954; doi: https://doi.org/10.1101/2020.10.19.343954, https://www.biorxiv.org/content/10.1101/2020.10.19.343954v1
- Peer reviewed and published scientific report.
Fiege, Jessica K., Joshua M. Thiede, Hezkiel Arya Nanda, William E. Matchett, Patrick J. Moore, Noe Rico Montanari, Beth K. Thielen, et al. 2021. “Single Cell Resolution of SARS-CoV-2 Tropism, Antiviral Responses, and Susceptibility to Therapies in Primary Human Airway Epithelium.” Edited by Kanta Subbarao. PLOS Pathogens 17 (1): e1009292. https://doi.org/10.1371/journal.ppat.1009292. https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1009292.