Researchers from the United States and the United Kingdom have shown that chronic lung disease is a risk factor for more severe COVID-19 and worse outcomes. A recent study published on the preprint server bioRxiv* in October 2020 shows that in patients with chronic lung disease, the pattern of gene expression in the different types of cell changes such that the lung epithelium is prepared to tackle SARS-CoV-2 infection. Also, the altered gene programs have an impact on the immune response.
In a large percentage of COVID-19 patients, the manifestations of the disease are nil to mild. Still, in others, it progresses to overwhelming illness, with acute respiratory distress syndrome (ARDS), multi-organ dysfunction, and even death. Some risk factors for disease progression have been identified, including advanced age, male sex, black and Asian ethnicity, and the presence of chronic health conditions such as high blood pressure, diabetes, and obesity.
Chronic lung disease, including chronic obstructive pulmonary disease (COPD) and interstitial lung disease (ILDs), is associated with a five-fold increase in the risk of severe COVID-19. ILD is also linked to a four-fold higher mortality following SARS-CoV-2 infection. COVID-19 outcomes are also especially poor in idiopathic pulmonary fibrosis (IPF).
Altered Basal Gene Expression Profile
The current study aims to explore the molecular mechanism of increased morbidity and fatality with SARS-CoV-2 infection. The researchers used single-cell RNA-sequencing (scRNAseq) datasets, along with 79 controls and 131 samples of chronic lung disease.
They found that in chronic lung disease, the baseline expression of those genes in specific cell types related to viral replication and the host immune response is altered, leading to dysregulation. This was seen in cells from the diseased lung epithelium.
They also found that immune cells from diseased tissue showed an exhaustion profile, while inflammatory genes were also expressed differently. Thus, the molecular processes in chronic lung disease independently confer increased risk for severe COVID-19 and of worse outcomes. Secondly, they found many entry factors expressed at a higher level in cells taken from diseased lungs, pushing up the viral entry score.
ACE2/TMPRSS2 Expression in Diseased Lung
SARS-CoV-2 entry depends on the cell receptor ACE2, and perhaps BSG and HSPA5, and the proteases required for priming, including TMPRSS2, CTSL, and FURIN. These molecules are expressed mainly on the upper and lower airways.
ACE2 is expressed at high levels in the nose, some AT2 cells, and absorptive cells in the gut. In the current study, ACE2 expression was confirmed to follow the same pattern, and the number of ACE2 cells was highest in AT2 cells. Previously, the expression of these factors has been examined only in asthma and COPD.
Both ACE2 and TMPRSS2 are upregulated in epithelial cells, but the number of ACE2 expressing cells of all types shows no significant increase in patients with COVID-19 compared to samples. In particular, specific cell types of a more proximal type are seen in the alveoli, while ACE2 expression is high in all lung tissue, but especially in the airway cells.
This may be due to an altered cellular composition of the lung epithelium in the patient with chronic lung disease, which causes the percentage of ACE2-expressing cells in the distal lung cells to rise, even while overall, lung tissue continues to show a low overall frequency of ACE2 cells and ACE2 protein. The former is, of course, thought to confer increased susceptibility to entry to SARS-CoV-2.
The current study shows that in all kinds of chronic lung disease, the same dysregulated pattern is obtained concerning these receptors/priming proteases in patients with COVID-19 and those without it. Overall, a composite score for the viral entry genes related to SARS-CoV-2 was increased in epithelial cell types from patients with chronic lung disease.
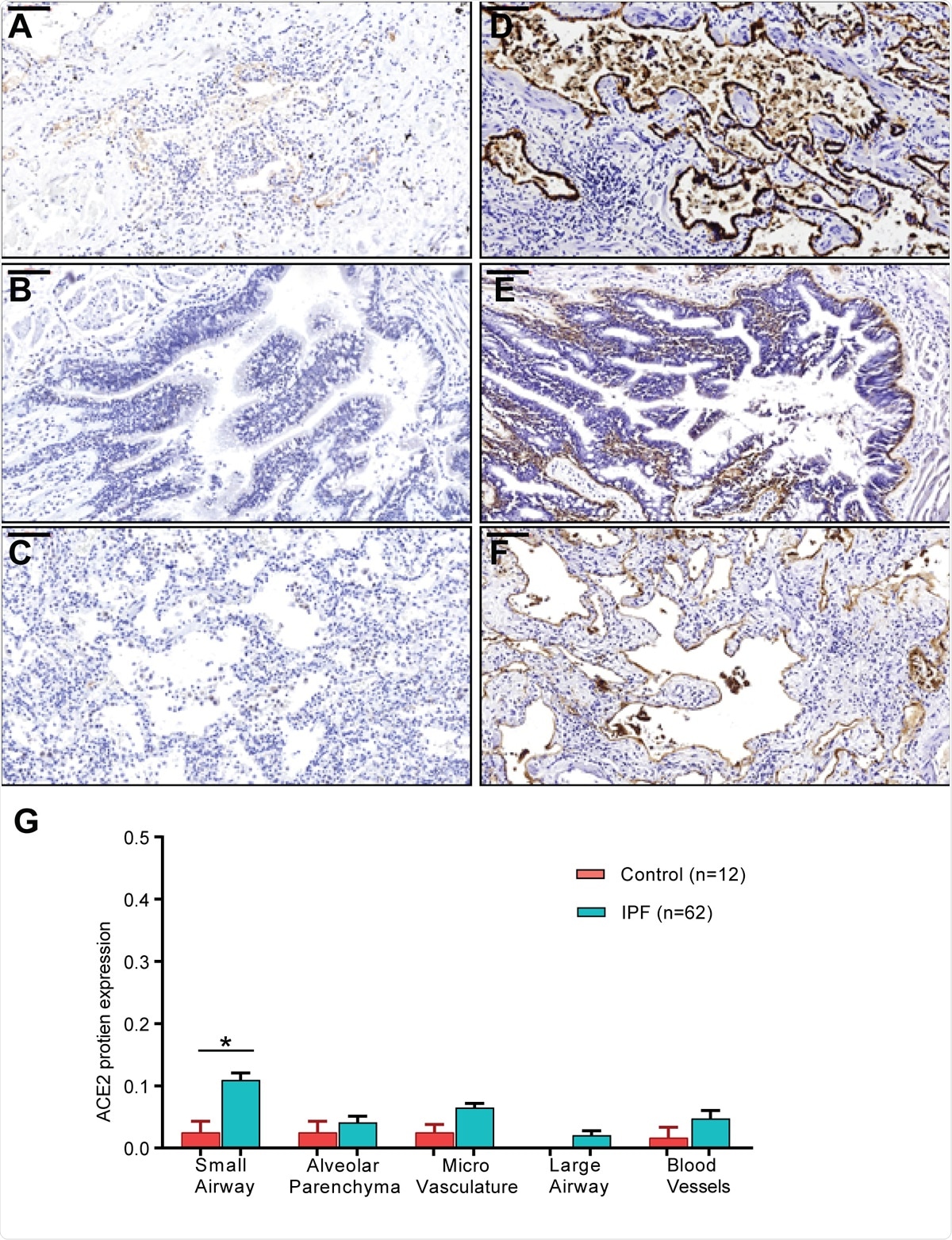
ACE2 and ITGB6 protein expression in IPF lung sections. (A-C) IPF lung sections stained for ACE2: (A) small airway, (B) large airway and (C) lung parenchyma. (D-F): IPF lung sections stained for αvβ6: (D) small airway, (E) large airway, and (F) lung parenchyma. (G) semi-quantitative evaluation of ACE2 scoring among control and IPF sections (both the percentage of staining and staining intensity of ACE2 expression;0-Negative; 1-0–⩽10%; 2-11–⩽25%; 3- ⩽26%). Significant differences between IPF and control were calculated using the Tukey HSD test, p-value < 0.05 *. Scale bar = 100μm.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
AT2 Cells Show Disease-Specific ACE2-Linked Gene Profiles
Again, they found that the ACE2 expression was correlated with a gene profile for each group of diagnoses, including those involved in antiviral defenses and immune regulation. Another finding was the increase in ACE2 protein expression in sections of airway tissue from patients with IPF. And finally, the type and number of immune cells in samples from patients with chronic lung disease were different from those of samples.
These findings make it doubtful that the primary contributor to the higher rates of severe disease and death in these patients is due to a higher risk of infection. Instead, the researchers suggest, “IPF lungs exhibit an abnormal expansion of epithelial cell programs.”
Dysregulation of Immunity Causes Increased Severity
Dysregulation of the immune response as a whole may underlie the worse outcomes in patients with chronic lung disease since it fails to achieve either adequate clearance of the virus or protection of the host against deleterious hyperinflammatory processes. In fact, these patients show elevated levels of inflammatory signaling molecules, including IL-2R, IL-6, IL-8, IL-10, and TNFα.
The researchers found that αvβ6 integrin expression is also increased. The increased expression of αvβ6 integrin in IPF patients is linked to poor outcomes. Being key to the activation of transforming growth factor (TGF)-beta, this may account for greater severity of lung damage in COVID-19, including more intense fibrosis.
Diseased lung tissue showed higher frequencies of CD4, CD8, and NK cells compared to control lungs. The interferon pathway, the IL6 cytokine pathway, and the major histocompatibility complex (MHC) class II genes are also expressed at higher levels in several immune cell types found more commonly in samples from chronic lung disease patients.
The researchers also found that AT2 cells in COPD patients with COVID-19 had higher expression levels of genes related to the immune response. These, in turn, may be related to the abnormally high levels of GCSF, an immune cell-stimulating factor that may contribute to the over-exuberant inflammatory response in severe COVID-19.
Implications
The researchers suggest, “The presence of immune-associated genes in these gene correlation profiles suggests that in patients with chronic lung diseases, ACE2+ AT2 cells are conditioned and primed to express these genes to cope with viral infection.”
On the other hand, some of these genes make up in part a negative feedback loop, which modulates the response to cytokines such as those mentioned above.
Overall, therefore, the lung's immune microenvironment with both IPF and COPD is thought to be imbalanced at baseline, before COVID-19. Such variations emphasize the change in the inflammatory response in chronic lung disease when the patient is faced with the virus, predisposing towards more severe COVID-19 and a worse outcome.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources