As the search for effective severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) inhibitors continues, a recent study by Yale University School of Medicine researchers published on the preprint server bioRxiv* in October 2020 reports the inhibitory effect of the drug merofloxacin on frameshifting during SARS-CoV-2 replication. This is an important proof-of-concept that supports the potential for targeting −1 PRF as an effective antiviral therapy in this pandemic as well as against other beta-coronaviruses.
It encodes a polyprotein, which is then cleaved into 16 nonstructural proteins (nsp) by the action of two proteases. Many proteins on the second half of the polyprotein play a vital part in viral transcription and replication. This includes nsp12, an RNA-dependent RNA polymerase (RdRp), RNA helicase (Hel/nsp13), and a proofreading exoribonuclease, among others.
The translation of ORF1b, the 3' side of ORF1ab, requires a frameshift to occur in all coronaviruses. When the ribosomes reach the end of ORF1a, some of them need to move in the retrograde direction by one nucleotide. This is the -1 programmed ribosomal frameshift (-1 PRF).
Following this frameshift, they continue to translate the rest of the ORF in the new -1 reading frame until the whole large polyprotein has been translated. If not, they will soon arrive at a stop codon in the 0 frame, which will end the translation prematurely. This will disable viral replication.
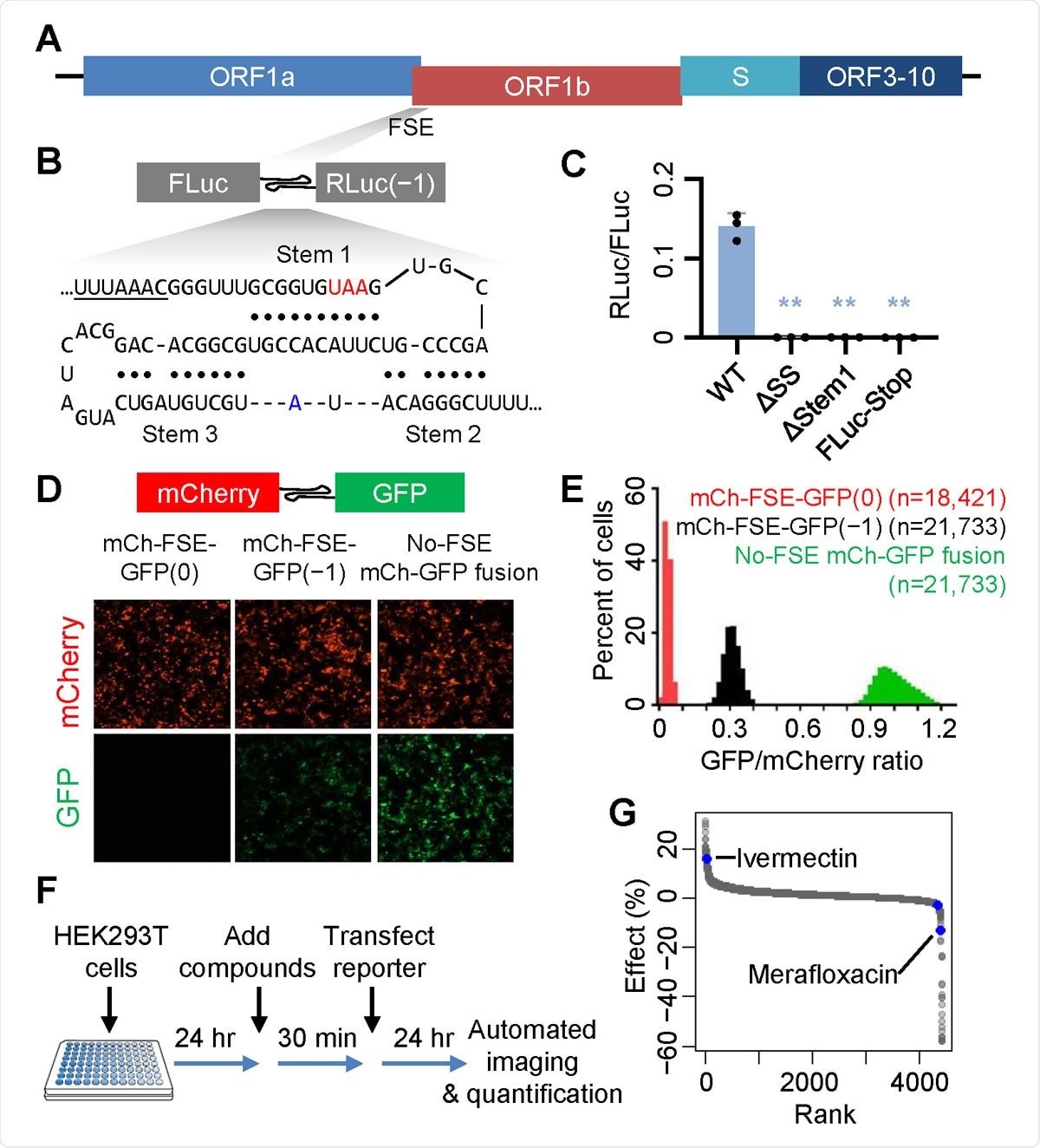
A high-throughput screen identifies SARS-CoV-2 −1 PRF modulators.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
FSE Structures
The −1 PRF region is often found to contain a 7-nucleotide slippery sequence (UUUAAAC in SARS-CoV-2) and a downstream stable secondary structure that functions as a frameshift-stimulating element (FSE). This latter brings about brief pauses in the ribosome's onward movement, allowing the tRNAs time to achieve the correct alignment with respect to the slippery sequence. This is partly required for the -1 Programmed Ribosomal Frameshifting (PRF).
One common FSE is the RNA pseudoknot. In SARS-CoV, SARS-CoV-2, and other betacoronaviruses, it is thought that perhaps a three-stem pseudoknot may act as the FSE. This structure is not very common in host mRNAs. It is, therefore often used to test the effect of explicitly disrupting viral gene expression, as by mutations and drugs that inhibit -1 PRF.
This has been demonstrated in HIV replication, both by an exogenous -1 PRF inhibitor and by clues indicating that it is part of the host's antiviral response.
Merofloxacin Inhibits SARS-CoV-2 Replication
In the current study, the researchers identified the fluoroquinolone antibiotic merofloxacin by high-throughput screening for −1 PRF inhibitors of SARS-CoV-2. They show that incubation with this compound suppressed SARS-CoV-2 replication in Vero cells.
Though ivermectin was also a -1 PRF inhibitor, it had cytotoxicity effects, unlike merofloxacin, which has only modest cytostatic effects even at high levels. It is a specific betacoronavirus -1 PRF inhibitor, unlike other fluoroquinolones that lack this activity. This suggests that the structural groups at certain specific sites may be responsible for the frameshift inhibition.
Since the FSE of the SARS-CoV sequence is almost identical to that of SARS-CoV-2, merofloxacin inhibits 1- frameshift in both almost equally. The half-maximal inhibitory concentration (IC50) is ~20 μM, which is superior to the 25% inhibition produced by another inhibitor called SHFL.
Merafloxacin is a specific inhibitor of betacoronavirus FSEs, but not of other coronaviruses or other common viruses. Other seasonal coronaviruses have a more elaborate pseudoknot structure, unlike the 3-stem pseudoknot of the betacoronaviruses.
Merofloxacin Inhibition Resists Escape Mutations
The researchers also found that merofloxacin-induced frameshift inhibition persists despite mutations in the viral genome. RNA viruses are known to have a rapid mutation rate, some of which may render the virus resistant to the antiviral agent. The study included mutations already known to exist in the virus.
They found very few mutations in the FSE sequence, which is to be expected in the light of the great importance of -1 PRF to viral replication. Among 20 mutations in this region, no less than 16 have occurred only once. Of the remaining four, two are in stem 1, and one each in stem 2 and 3.
Being on the stem, none of these could result in an alteration in the three-stem structure of the pseudoknot. However, the researchers found that -1 PRF inhibition by merofloxacin occurred with all these variants without loss of efficacy.
Proceeding from these already existing mutations that did not change the pseudoknot structure, they then explored the effect of other mutations that disturbed the structure. They found that these caused a reduction in frameshifting by 10% to 77%, depending on their disruption. One of the most significant reductions was caused by a mutation in single unpaired uridine, indicating that it has an unknown function. Both this mutation, U13485, and the following G13486 are completely conserved among human coronaviruses.
Though these mutations generate a wide range of structural perturbations, the inhibition of frameshift produced by merofloxacin acting on all these variants remained almost the same. This indicates that this action persists, and escape mutations are unlikely to emerge.
Frameshift inhibition impedes SARS-CoV-2 replication
The researchers then examined the effect of the inhibitor on viral replication. They found that merofloxacin inhibited replication from the lowest concentration of 1.25 μM, at which the half-maximal effective concentration (EC50) was 2.6 μM. When the drug concentration was raised to 40 μM or above, the virus became undetectable. The drop in viral yield was almost exponentially related to the -1 PRF inhibition. This indicates that the rate of replication of the virus depends on frameshift efficiency.
Implications and Future Directions
The FSEs of coronaviruses are highly efficient, and this study explores the measure to which this attribute relates to viral viability by using a newly identified frameshift inhibitor. The researchers describe the results: "The near-exponential relationship between the viral titer and −1 PRF efficiency points to a simple model in which ORF1b translation sets the limit of SARS-CoV-2 growth rate."
This justifies the development of −1 PRF inhibitors to effectively antagonize SARS-CoV-2 growth and other RNA viruses of the same type of PRF. More research remains to be carried out to elucidate the mechanism of inhibition of -1 PRF by merofloxacin.
Other Possible Explanations
The results could also be explained by the absolute need for highly efficient PRF to achieve the right stoichiometric correlation between the replication complex enzymes. The loss of this balance by reducing or increasing the efficiency of -1 PRF by even a little bit impacts viral growth severely.
Another possibility is that the small reductions in each of the translated rate-limiting enzymes of the replicase-transcriptase complex accumulate to cause a marked reduction in viral replication.
It could be due to direct binding of the FSE, disturbing the stability of the pseudoknot and thus decreasing the necessary pausing of the ribosome which is required for frameshifting. Another view is that this binding confers extra stability to the pseudoknot so that the arriving ribosomes are stuck too long, collide or have to queue, all disturbing the smooth translation elongation process.
Other possibilities include the stabilization of an incorrect FSE conformation, which precludes frameshifting. Such structures have been recently demonstrated in cells infected by the virus. If merofloxacin is found to interact with one or more of such unproductive structures, it could reduce the FSE efficiency by the resulting smaller fraction of productive RNAs.
Finally, merofloxacin may act on modulatory host factors that act on the reverse frameshift, such as host RNA helicases, which might participate in pseudoknot unfolding following the -1 PRF but prior to the continuation of translation. Fluoroquinolones are known to target bacterial DNA topoisomerases. Further study will reveal the analogous enzymes in higher organisms, including RNA analogs.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources