By leveraging nucleotide triphosphate analog-induced cell death, Chinese researchers unveiled genes that affect the import, export, and metabolism of pervasively used antiviral drugs in the treatment of coronavirus disease (COVID-19). This exciting study is currently available on the bioRxiv* preprint server.
Study: Genetic determinants of COVID-19 drug efficacy revealed by genome-wide CRISPR screens. Image Credit: CI Photos / Shutterstock
The global spread of COVID-19, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has paralyzed health systems and economies worldwide. As the quest for effective drugs and vaccines continues, scientists and healthcare workers turned towards drug repurposing efforts for treating this infection.
Some notable examples of drugs currently used in this combat against COVID-19 are immunomodulatory agents such as dexamethasone and colchicine, antiviral drugs remdesivir, ribavirin, and favipiravir, as well as antimalarial drugs chloroquine and hydroxychloroquine.
Nevertheless, due to the urgency and lack of relevant knowledge, most of these drugs have been applied with little consideration of genetic biomarkers. Considering a rapid disease progression and the availability of myriad treatment options, we need to discern genetic modulators for each COVID-19 drug so patients can be matched to the most effective drug choice.
The researchers – led by Dr. Wei Jiang from the Shanghai Institute of Biochemistry and Cell Biology and the University of Chinese Academy of Sciences – did just that by pursuing the first genome-wide CRISPR screen on the aforementioned COVID-19 drugs and revealed important data on both resistance and sensitization mechanisms.
Genome-wide libraries for genetic biomarkers
Since the lack of executable screen endpoints and lack of toxicity present two substantial hurdles for genome-wide screens when antiviral drugs are concerned, this group of scientists decided to use nucleotide triphosphate analog-induced cell death as a surrogate screen endpoint to overcome these obstacles.
"From a method standpoint, this screening strategy is the first that is capable of employing genome-wide libraries to identify genetic biomarkers for antiviral drugs," study authors explain their methodological approach.
In a nutshell, the researchers designed a screening approach that could utilize genome-wide single guide RNA libraries to discover genes indispensable for the tested drugs' action systematically. This approach is broadly applicable to many existing and emerging viral illnesses.
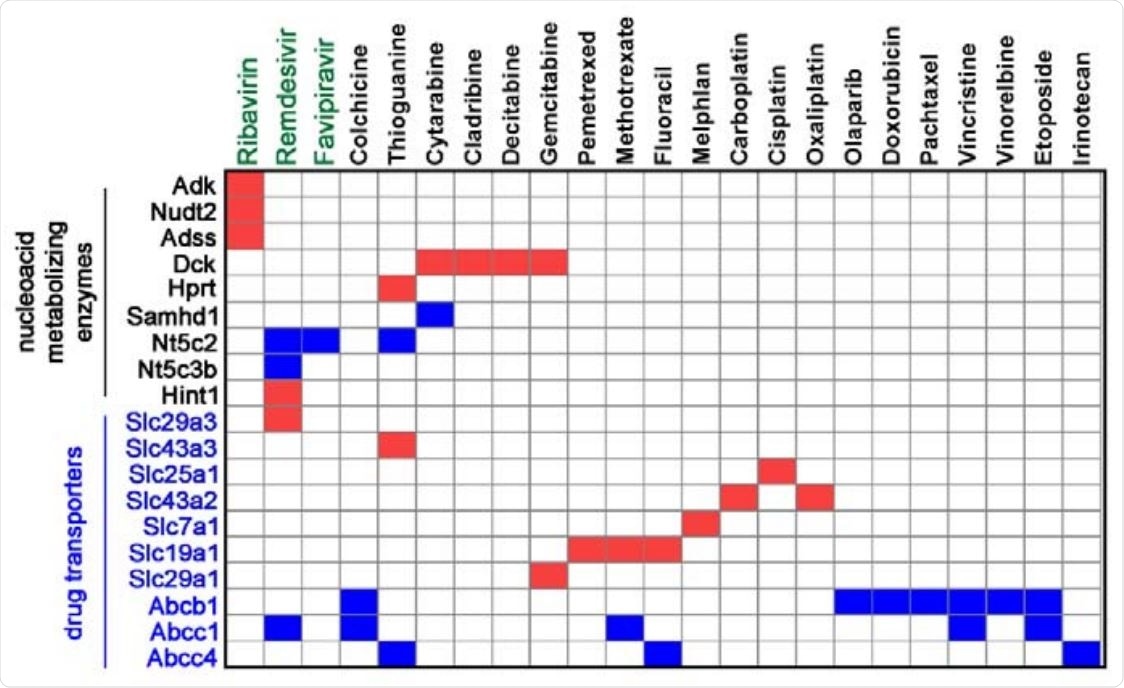
Summary and comparison of drug-metabolizing enzymes and transporters identified in genome-wide CRISPR screens using our platform. Red boxes indicate enrichment of sgRNA against a gene in corresponding drug screen, whereas blue boxes indicate depletion.
Finding genes crucial for drugs' action
The results of this study suggest that several gene groups that are involved in autophagy, but also lipid and cholesterol synthesis, may significantly modulate the action of chloroquine drugs. In turn, such a list of genes may be useful to appreciate and guide the clinical use of chloroquine drugs.
Furthermore, it was shown that alkaloid drug colchicine might not sufficiently suppress the immune response for symptom alleviation in many patients with a specific constellation of genes, and the study further shed light on exact cytotoxic mechanisms of microtubule poisons (i.e., a significant group of anticancer drugs whose death mechanisms are still unclear).
The top hit in the researcher's ribavirin screen was adenosine kinase – an enzyme that catalyzes adenosine diphosphate (ADP) formation from adenosine. This turns out to be the key to the antiviral activity of ribavirin.
Finally, the study demonstrated essential roles of SLC29A3 (encoding a nucleoside transporter protein) and HINT1 (coding for homodimeric purine phosphoramides) in the importation and activation of remdesivir, while drug-transporting gene ABCC1 and dephosphorylation enzyme gene NT5C2 were unveiled as pivotal in exporting and inactivating the same drug.
Advancing our treatment approach
"Transportation, activating and detoxifying mechanisms uncovered in this screen may point to clinical biomarkers of COVID-19 drugs", say study researchers. "If such knowledge could be established in the future, it may help better understand the efficacy of individual drugs in the context of COVID-19", they add.
The results available in this bioRxiv paper also suggest that infected individuals with specific genetic variation may respond differently to remdesivir and favipiravir, but not to ribavirin. Moreover, medically relevant single nucleotide polymorphisms for many of the genes identified in this screen have been reported in the human population.
In conclusion, all the screens delineated in this study will help advance the understanding of the exact action of COVID-19 drugs, potentially revealing useful genetic biomarkers for highly targeted disease management.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Jiang, W. et al. (2020). Genetic determinants of COVID-19 drug efficacy revealed by genome-wide CRISPR screens. bioRxiv. https://doi.org/10.1101/2020.10.26.356279, https://www.biorxiv.org/content/10.1101/2020.10.26.356279v1
- Peer reviewed and published scientific report.
Wei, Jin, Mia Madel Alfajaro, Peter C. DeWeirdt, Ruth E. Hanna, William J. Lu-Culligan, Wesley L. Cai, Madison S. Strine, et al. 2021. “Genome-Wide CRISPR Screens Reveal Host Factors Critical for SARS-CoV-2 Infection.” Cell 184 (1): 76-91.e13. https://doi.org/10.1016/j.cell.2020.10.028. https://www.cell.com/cell/fulltext/S0092-8674(20)31392-1.