As seroprevalence studies continue across the globe to estimate the true extent of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and the world's progress towards herd immunity, a new study published on the preprint server medRxiv* in October 2020 reports that there are large differences in the viral load in different communities. Moreover, the authors say that the cycle threshold values required to obtain a positive result for viral RNA by polymerase chain reaction may provide an early warning of an outbreak.
As the COVID-19 rates continue to mount, after an initial fall, many governments are exploring ways to detect and contain new infections. Lockdowns and mass testing are often very taxing in terms of immense economic and social impact. Large-scale asymptomatic testing in a low-risk population could yield a large or significant share of false positives, limited only by the test's specificity.
As the specificity of the test goes down, the positive predictive value (PPV) of the result declines even with a 100% sensitive test. This could throw up large numbers of false positives, depending on the prevalence. For instance, with a 0.1% prevalence in a community of 100,000, a 100% sensitive test and 99.99% specific could throw up all 100 true positives, but also 100 false positives.
Modeling also examines the effectiveness of many potential control plans. However, this uses existing infectivity estimates. The need is for independent community-level surveillance studies carried out on both symptomatic and asymptomatic cases.
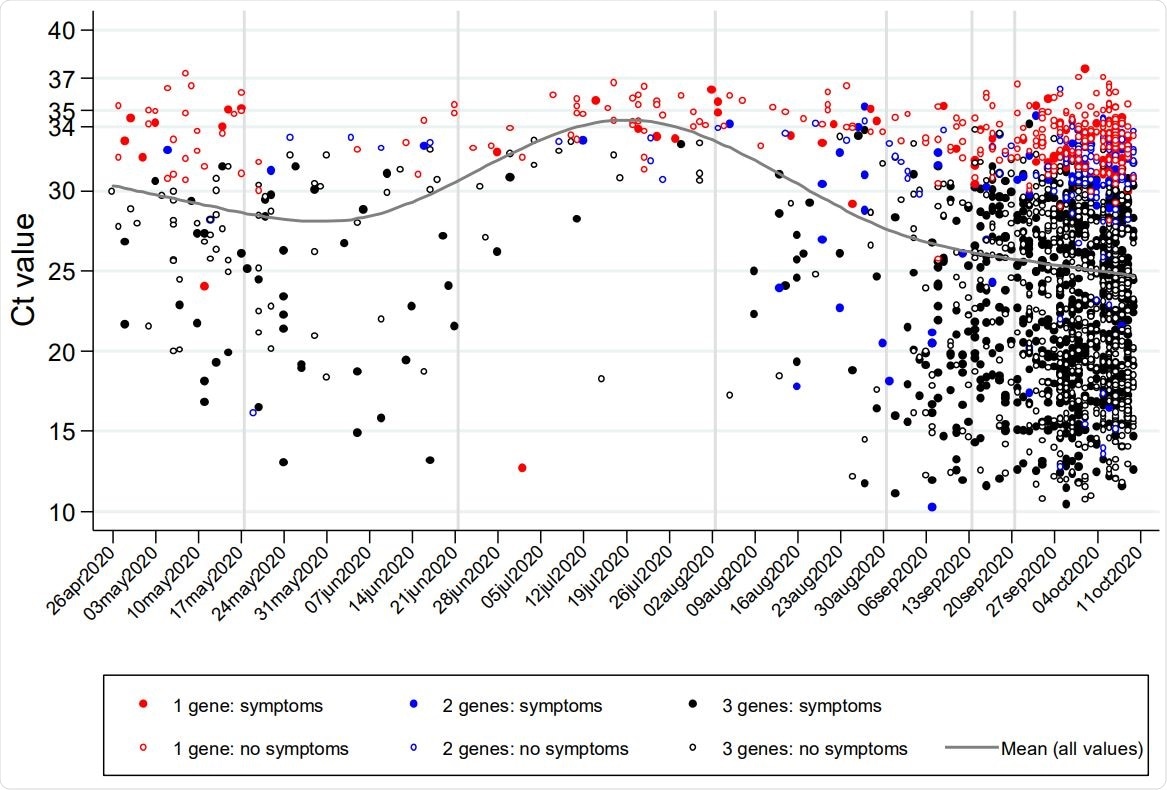
Variation over calendar time in Ct values (raw values (A) and distribution (B)), reported symptoms (C), and supporting evidence (D) for positive RT-PCR tests from community surveillance

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Asymptomatic Infection Could Drive Resurgence
The true level of asymptomatic infection is also unknown, is estimated at below a fifth of all infections. However, this figure is derived from many studies of contacts of patients known to be infected. Community-based programs show a higher prevalence of asymptomatic infection.
Earlier research suggests that asymptomatic infection is associated with a lower transmission rate, perhaps due to a lower viral load since this is among the most important factors influencing transmission. However, estimating the mean proportion of asymptomatic infection without accounting for the patient characteristics and the viral load ignores time-related changes in these vital factors.
Viral Characteristics
The current study explores variations in patients with COVID-19 who have been confirmed by testing from the first five months of the UK COVID-19 Infection Survey (CIS). This was an extensive longitudinal follow-up of households representing the general population. There were ~233,000 subjects, with ~844,000 nose and throat swabs, a median of 3 per individual tested. About a quarter had only been tested once.
The total number of positive individuals was ~1,500, or 0.2% of the total swabs. About 40% were positive at first contact, and 60% became positive later. The median number of swabs in the latter group before a positive result was 2.
The number of genes that tested positive was one, two, or three for ~20%, 10%, and 70%, respectively. In only 4 cases was the Ct value over 37. The median Ct was ~26, and Ct values showed excellent correlation with the increasing number of detected genes, whatever the exact gene detected.
Evidence Supporting Positive Results
Of all the positive tests, ~80% were highly in favor of true positivity (two or three genes detected), with ~10% and 6% showing moderate or low evidence (one gene detected), respectively. The former was more likely to be symptomatic and to belong to people in high-risk occupations or people who came from households with confirmed infections.
Factors Determining Ct values
The researchers found that Ct values were lower, indicating higher viral loads, as the number of genes detected increased. When all three genes were positive, the Ct was ~10 lower, compared to with a single positive gene. Ct values were also decreased, though by lower margins, if patients were symptomatic at the time of testing, by ~2.
There was a trend towards lower Cts in patients with fever, cough, or anosmia vs. other symptoms, by ~2. However, sex, ethnic origin, and age, as well as poverty, showed no association.
Finally, Ct values were ~2 lower if the test came from an individual from a household with confirmed infection. Ct values were always higher at the time of the first positive test and in the poorest patients, at ~3 lower, as well as when the positive was not the first test but a subsequent test, at ~1.4 lower.
Almost all positive tests for only one gene who had no symptoms had Ct above 30, but Ct showed much more variations among asymptomatic patients with all three genes positive. The chances of symptomatic infection were in direct inverse proportion to Ct values from 25 to 35, but showed no change below a Ct of 25.
Ct Values and Symptomatic Infections Change with Time
The time of testing also had a significant effect on the test results. In July or August, the number of positives was much less when the Ct was below 30, while symptomatic infection was far less common at the time of a positive test. The supporting evidence was also likely to be weaker during this period.
However, this should be balanced by the observation that positive patients with stronger evidence also showed higher Ct, both earlier and later. Again, more tests resulted in positives with low evidence during April/May and this period, when only S genes were detected. Later, all three classes of evidence increased in the same proportion.
Seroconversion and Viral RNA Testing
Very few antibody tests were done (6%), and before-after comparisons could be made in very rare cases. Over three-quarters of those with a positive first swab who had a preceding antibody test were negative. However, seven patients became positive even with high anti-spike antibodies before the PCR test was done.
Future studies will explore whether this is due to reinfection, in which case it could be related to continuing viral transmission even in the presence of specific antibodies.
Implications and Future Directions
The researchers concluded that viral load (denoted by Ct values) varied largely in the community. The greatest relationship was between symptomatic status at the time of the test and the number of positive viral genes. Even with all three genes being detected, but without symptoms, the Ct showed marked differences. Such individuals, with a low Ct and, therefore a high viral load, but without symptoms, may play a large role in super-spreading events.
The Ct values were mostly corroborated by finding two or three positive genes per positive test (higher evidence). This shows that false positives probably did not skew the results of the study. There were more mild infections at this time, with a false positive rate of ~0.005%.
The study also showed the temporal dynamics of Ct values, which indicates that the viral burden keeps changing in infected individuals. The clinical features appeared less severe, and the Ct values higher, in July/August than later on, which seems to contradict the theory that viral fitness is less now than at the onset of the pandemic. However, this could also, perhaps, be explained by changes in behavior, climate or environment, which affected viral transmission.
The study concludes, “Our findings support the use of Ct values and genes detected more broadly in public testing programs, predominantly testing symptomatic individuals and case contacts, as an “early warning” system for shifts in potential infectious load and hence transmission.”
Recent work has shown that Ct levels were lower than before in August, but only in September did the rates of positive tests begin to rise. The Ct data, universally stored on laboratory servers, could thus be put to good use, alongside symptom status and risk factor assessment, to shape optimal responses on both individual and public health fronts.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Walker, A. S. et al. (2020). Viral Load In Community SARS-Cov-2 Cases Varies Widely and Temporally. medRxiv preprint. doi: https://doi.org/10.1101/2020.10.25.20219048. https://www.medrxiv.org/content/10.1101/2020.10.25.20219048v1
- Peer reviewed and published scientific report.
Walker, A Sarah, Emma Pritchard, Thomas House, Julie V Robotham, Paul J Birrell, Iain Bell, John I Bell, et al. 2021. “Ct Threshold Values, a Proxy for Viral Load in Community SARS-CoV-2 Cases, Demonstrate Wide Variation across Populations and over Time.” Edited by M Dawn Teare and Miles P Davenport. ELife 10 (July): e64683. https://doi.org/10.7554/eLife.64683. https://elifesciences.org/articles/64683.