The COVID-19 pandemic, which continues to spread worldwide, is caused by the SARS-CoV-2 pathogen. It belongs to the betacoronavirus group - along with severe acute respiratory syndrome coronavirus 1 (SARS) and Middle East respiratory syndrome (MERS) - and is a positive-sense single-stranded RNA virus. Its genome sequence is about 30,000 nucleotides long, containing 14 open reading frames (ORFs). The two overlapping 5’-end frames encode large polypeptides that are cleaved by viral proteases Nsp3 and Nsp5, generating the unstructured proteins Nsp1 to Nsp16.
Studies have reported that Nsp1, which is the first protein produced by a virus, is a critical component that affects the activity of the infected cell and the development of viral particles. It is also responsible for suppressing host interferon response, one main mechanism of defense against the virus.
The SARS-CoV-2 Nsp1 is a small protein with 180 amino acid residues and N- and C-terminals. The N-domain is responsible for the efficient translation of the virus. The C-domain binds to the entry channel of the 40S ribosome unit, which prevents the translation of the host cell messenger RNA (mRNA). The SARS-CoV-2 Nsp1 is also found in 80S ribosomes.
A previous study has shown these 80S ribosomes also have proteins involved in translation termination, the final stage of protein biosynthesis where polypeptides are released from the ribosome. Translation termination is important because suppressing this process prevents further translation.
How Nsp1 affects translation termination
The researchers investigated the role of SARS-CoV-2 Nsp1 in translation termination in host cells and reported it in a paper published in the bioRxiv* server.
For their study, the researchers tested SARS-CoV-2 Nsp1 – expressed in E. coli BL21 cells – on translation in rabbit reticulocyte lysate (RRL). They found that Nsp1 stimulates translation termination. When Nsp1 was added to a pre-termination complex, along with a release factor eRF1, it increased the amount of termination complex formed. They determined that Nsp1 stimulates translation termination before peptide release.
By analyzing different Nsp1 mutants, the team found that the Nsp1 C-domain is involved in translation termination, and the N-domain is responsible for its activation in translation termination. Moreover, they also found that the full protein is required for translation termination.
The authors also discovered that Nsp1 increased the readthrough efficiency in the premature termination codon (PTC) for UAG and UGA codons, which meant a decreased termination of translation. The effect of PTC readthrough is controlled by C-domain binding of Nsp1 to 40S. Using a model mRNA system with three PTC stop codons, the authors found that Nsp1 stimulated the formation of termination complex in the presence of an eRF1-eRF3-GTP complex.
Based on their results, the team suggests that Nsp1 stimulates translation termination during stop codon recognition by its N-terminal domain. To prevent the translation of mRNAs that have already passed the translation stage, “it would be reasonable to stimulate termination at the stop codons and remove the release factors and 40S ribosome subunits from the pool of active components,” write the authors. It’s likely Nsp1 plays a role in this process. Thus, Nsp1 can prevent host initiation of translation by binding to the 40S ribosome units.
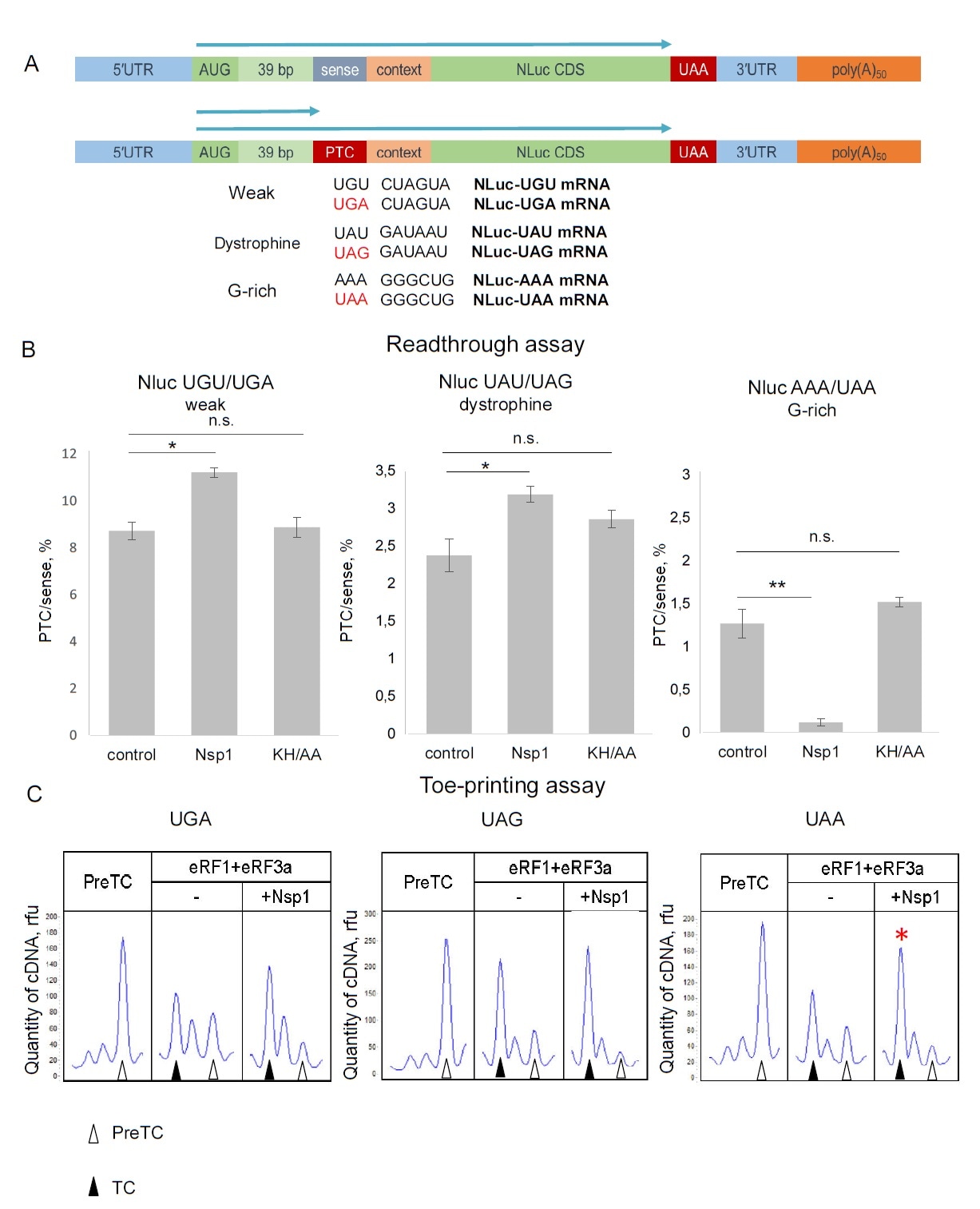
Nsp1 affected PTC readthrough in RRL. (?) Scheme of the NLuc constructions containing the leaking contexts in ß-globine 5’ UTR followed by sense codon or PTC. (B) In vitro translation in RRL with Nluc mRNA containing sense codons or PTCs in the presence/absence Nsp1 and Nsp1 mutant - KH/AA, relative luminescence units (rlu). n=4, 3, 3, respectively, mean±SE, * - p<0.05, ** - p<0.01. (C) An example of raw toe-printing data. TC formation was induced by the addition of eRF1+eRF3a in presence of Nsp1 on different stop codons. The TC corresponds to the black triangle, and the preTC corresponds to the white triangle. Red stars indicate the increased quantity of ribosomal complexes, shifted from the preTC to the TC state.
Drugs that disrupt Nsp1 functioning
The team also studied the effect of two drugs, which affect different parts of the termination complex, on the Nsp1 activation of translation termination: gentamicin and mefloquine. The antibiotic gentamicin is known to stimulate PTC readthrough and binding to 80S ribosome. Mefloquine, an antimalarial, is observed to bind to the 80S ribosome of a malaria parasite. Hence, it is possible that these drugs affect translation termination, competing with Nsp1.
The researchers tested the effect of these drugs on peptide release and termination complex formation. Neither drug had any effect in the presence of release factors eRF1 and eRF3a, but in the presence of Nsp1, gentamicin stimulated peptide release. However, mefloquine in the presence of Nsp1 decreased peptidyl-tRNA hydrolysis. Further tests showed that gentamicin and mefloquine affect peptide release in the presence of Nsp1 differently. This could be because the two drugs affect different regions of the 80S ribosome and so modify different steps of the translation termination process.
Because Nsp1 affects translation termination by stimulating stop codon recognition, drugs that induce PTC readthrough could potentially curb infection. The authors write that the use of mefloquine as a potential antiviral against COVID-19 could be because it disrupts Nsp1 function during termination. They suggest further studies into this antimalarial’s potential to combat SARS-CoV-2 as well as other possible drugs that have the potential to disrupt Nsp1’s function.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Shuvalov, A. et al. (2020) Nsp1 of SARS-CoV-2 Stimulates Host Translation Termination. bioRxiv. https://doi.org/10.1101/2020.11.11.377739, https://www.biorxiv.org/content/10.1101/2020.11.11.377739v1
- Peer reviewed and published scientific report.
Shuvalov, Alexey, Ekaterina Shuvalova, Nikita Biziaev, Elizaveta Sokolova, Konstantin Evmenov, Nikolay Pustogarov, Aleksandra Arnautova, Vera Matrosova, Tatiana Egorova, and Elena Alkalaeva. 2021. “Nsp1 of SARS-CoV-2 Stimulates Host Translation Termination.” RNA Biology 18 (sup2): 804–17. https://doi.org/10.1080/15476286.2021.1999103. https://www.tandfonline.com/doi/full/10.1080/15476286.2021.1999103.