A team of researchers in Europe has created a comprehensive data resource to help improve understanding of how coronavirus disease 2019 (COVID-19) progresses and how biomarkers could be used to guide treatment approaches and disease monitoring.
The team from Germany, the UK, and Austria – characterized the time-dependent progression of COVID-19 using diagnostic and proteomic markers at more than 600 hundred sampling points among a cohort of hospitalized patients.
The study revealed that an initial surge in the inflammatory response to infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – the agent that causes COVID-19 – is a key determinant of disease trajectory.
The team also showed that routine diagnostic parameters and early proteomic measurements could be used to predict the future progression of the disease.
The researchers accurately predicted survival among severely ill patients using results from proteome samples that had been recorded weeks before clinical outcomes were reached.
They also identified age-specific molecular responses to the illness that contribute to increased inflammation among older patients.
"Our study provides a deep and time-resolved molecular characterization of COVID-19 disease progression and reports biomarkers for risk-adapted treatment strategies and molecular disease monitoring," writes Florian Kurth (Charité – Universitätsmedizin Berlin) and colleagues.
A pre-print version of the paper is available on the server medRxiv*, while the article undergoes peer review.
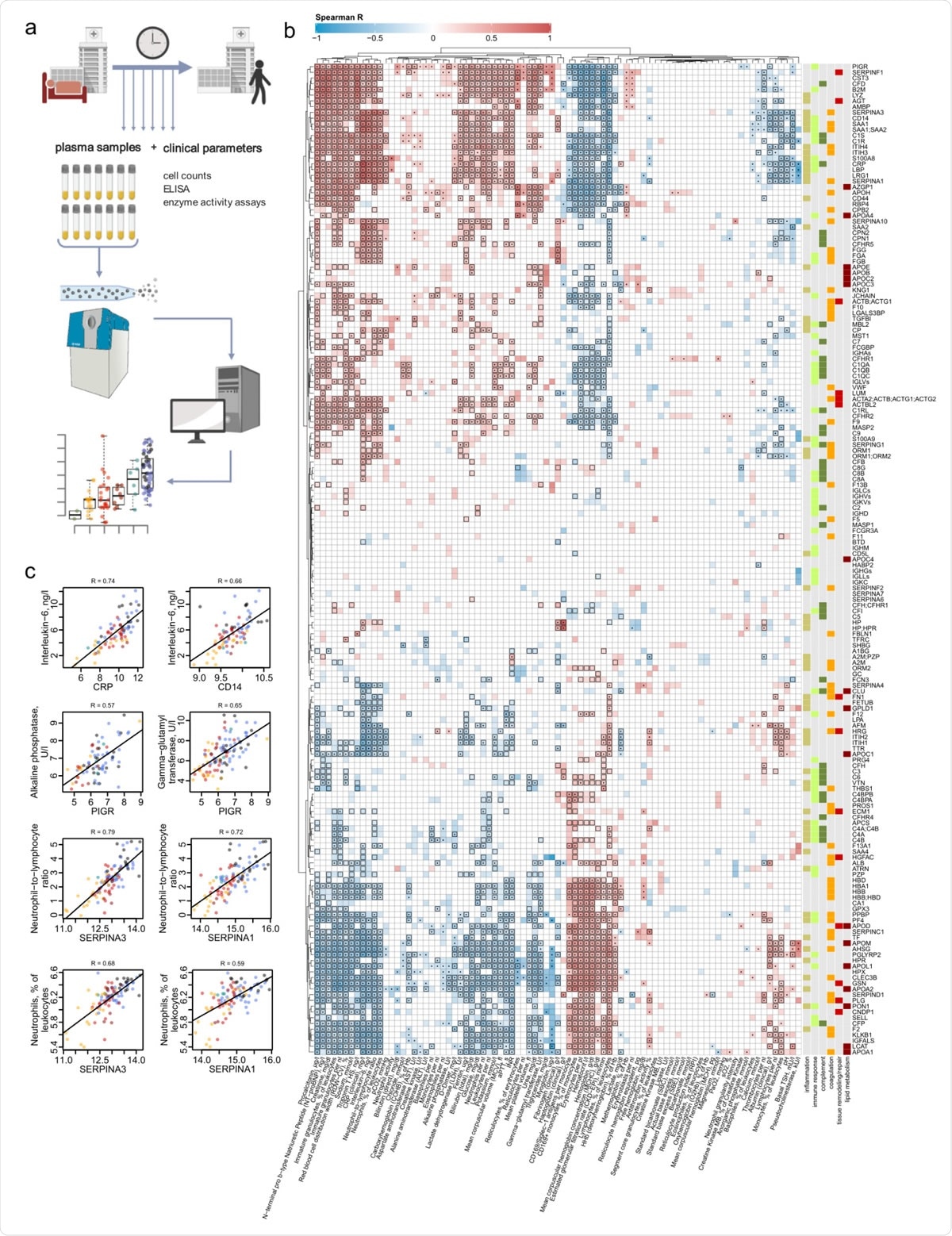
Interdependence of clinical, diagnostic, physiological and proteomic parameters in COVID-19. a. Study design. Schematic of the cohort of 139 patients with PCR confirmed SARS-CoV-2 infection treated at Charité University Hospital Berlin. Plasma proteomics and accredited diagnostic tests were applied at 687 sampling points, to generate high-resolution time series data for 86 routine diagnostic parameters and 309 protein quantities b. Covariation map for plasma proteins and routine diagnostic and physiological parameters (log2-transformed). Statistically significant correlations (Spearman; P < 0.05) are coloured. Dots indicate statistical significance after row-wise multiple-testing correction (FDR < 0.05), black rectangles – column-wise.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Highly variable disease presentation makes prognosis challenging
The clinical presentation of COVID-19 is highly variable, ranging from mild or even no symptoms to severe disease, organ failure, and death.
Accurate predictive markers of disease trajectory are urgently needed to help guide resource allocation, monitor therapies' effectiveness, and personalize treatment strategies.
"Patients likely to suffer progression to severe disease and organ failure and those likely to remain stable could be identified early, which is particularly valuable in scenarios where health care systems reach capacity limits," said Kurth and colleagues. "Prognostic panels would also optimize the monitoring of novel treatments, thereby accelerating clinical trials."
Where does proteomics come in?
Proteomics studies have shown that the complement cascade and the acute phase response are activated in response to COVID-19 and that proteomic and diagnostic parameters can point to the underlying mechanisms involved and potential treatment approaches.
However, it is unclear how factors influencing the host response change over time and as the disease progresses or how these factors are interrelated during pathological processes.
What did the researchers do?
The team conducted a time-resolved deep clinical and molecular phenotyping during different stages of disease among 139 patients hospitalized with COVID-19.
The researchers measured 86 accredited routine clinical diagnostic markers and plasma proteomes at 687 sampling points to obtain a detailed picture of how the molecular phenotype changes over time.
They studied the associations between these different markers and how their levels changed to distinguish between the different trajectories that characterize mild and severe disease.
What did the study find?
The patients' molecular phenotypes reflected an initial spike in the systemic inflammatory response to COVID-19 and a gradual alleviation of most markers of disease severity over time, suggesting an eventual return to the baseline phenotype.
This alleviation was followed by a protein signature indicative of tissue repair, modulation of the immune response and metabolic reconstitution.
The inflammatory host response during the early stage of disease was found to be a major determinant of clinical progression and revealed proteomic and diagnostic markers that could classify the requirement for supplemental oxygen and mechanical ventilation.
These prognostic markers correlated with the remainder of time spent in hospital and could predict the time to recovery among patients with mild disease.
The study also revealed markers that could predict clinical worsening across all stages of disease severity.
Furthermore, among severely ill patients, the molecular phenotype of the host response during the early disease stage accurately predicted survival weeks before the clinical outcome was reached (average time 39 days).
The results also showed proteomic signatures of higher basal inflammation with increasing age, which may partly explain the increased risk of severe disease among older individuals, suggest the researchers.
"Whereas several approaches of targeted anti-inflammatory treatment have not been successful to prevent clinical deterioration in COVID-19 so far, our study indicates that this special patient population might benefit particularly from treatments that mitigate the early inflammatory host response," say Kurth and colleagues.
"A rich data resource for understanding the progression of COVID-19"
"We have created a rich data resource for understanding the progression of COVID-19 until outcome," write the researchers.
Although prognostic assessments based on measurements taken at single time points may help guide the timely management of patients and resource allocation, measuring dynamic parameters over time can be particularly valuable for motoring treatment, including the experimental therapies being used in clinical trials, says the team.
"Our study provides comprehensive information about the key determinants of the varying COVID-19 disease trajectories as well as marker panels for early prognosis that can be exploited for clinical decision making, to devise personalized therapies, as well as for monitoring the development of much needed COVID-19 treatments," conclude Kurth and colleagues.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Kurth F, et al. A time-resolved proteomic and diagnostic map characterizes COVID-19 disease progression and predicts outcome. medRxiv, 2020. doi: https://doi.org/10.1101/2020.11.09.20228015, https://www.medrxiv.org/content/10.1101/2020.11.09.20228015v1
- Peer reviewed and published scientific report.
Demichev, Vadim, Pinkus Tober-Lau, Oliver Lemke, Tatiana Nazarenko, Charlotte Thibeault, Harry Whitwell, Annika Röhl, et al. 2021. “A Time-Resolved Proteomic and Prognostic Map of COVID-19.” Cell Systems 12 (8): 780-794.e7. https://doi.org/10.1016/j.cels.2021.05.005. https://www.cell.com/cell-systems/fulltext/S2405-4712(21)00160-5.