Coronavirus disease (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in late 2019 in Wuhan, China, and rapidly progressed into a devastating pandemic affecting global public health and crippling economies. So far, the COVID-19 pandemic has caused more than 55.4 million infections and over 1.33 million deaths worldwide.
COVID-19 targets the upper and lower respiratory systems and causes flu-like symptoms in most infected people. Although many COVID-19 patients experience only mild symptoms, some patients have severe symptoms leading to massive lung damage. Treatment options for COVID-19 are limited and the crude mortality rate estimated by the WHO is around 2.9%. Although a preventive vaccine for COVID-19 could eventually become available, unless sufficient herd immunity is achieved, COVID-19 could potentially cause significant morbidity and mortality over the coming years.
This highlights the significance of understanding the role of the immune system in the progression and clinical outcome of COVID-19 patients to improve clinical management and develop effective vaccines and therapeutic interventions. A better understanding of the role of immunity might help identify applicable biomarkers that can predict the clinical outcome of COVID-19.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
IgM, IgG, and IgA antibodies increase and stay elevated during COVID-19 progression
SARS-CoV-2, like SARS-CoV and MERS-CoV, is part of the betacoronavirus family and its genome encodes 4 major structural proteins - envelope (E), spike (S), membrane (M), and nucleocapsid (N); 15 non-structural proteins - Nsp1-10 and Nsp12-16; and 9 accessory proteins. The S protein has the N-terminal S1 peptide with a key receptor-binding domain region and C-terminal S2 fragment. It plays an important role in viral attachment, fusion, and entry into the host cells with the viral receptor angiotensin-converting enzyme 2.
Rapidly growing serological evidence shows that IgM, IgG, and IgA antibodies against the S or N proteins evolve rapidly in the serum of asymptomatic as well as symptomatic COVID-19 patients within a week of infection or symptom onset and stay elevated with progressing disease. However, not much is known about humoral immune responses to the rest of the structural and non-structural SARS-CoV-2 proteins during disease progression.
Novel proteome microarray with 20 SARS-CoV-2 proteins to understand IgM/IgG responses
Researchers from the Huazhong University of Science and Technology and Shanghai Jiao Tong University, China, recently built a proteome microarray with 20 out of the 28 predicted SARS-CoV-2 proteins to help understand IgM/IgG responses specific to SARS-CoV-2. Their study is published on the preprint server medRxiv* prior to undergoing the peer review process.
The team assumed that anti-SARS-CoV-2 IgM and IgG antibodies may serve as biomarkers that can predict prognosis and outcome in COVID-19 patients. Using the SARS-CoV-2 proteome microarray, they analyzed IgM/IgG responses in 1,034 hospitalized COVID-19 patients against 20 SARS-CoV-2 proteins. The analysis was performed on admission and continued for 66 days. The results from the microarray were correlated with laboratory test results, clinical information, and patient outcomes. They used the Cox proportional hazards model to determine the association between SARS-CoV-2-specific antibodies and mortality in COVID-19 patients.
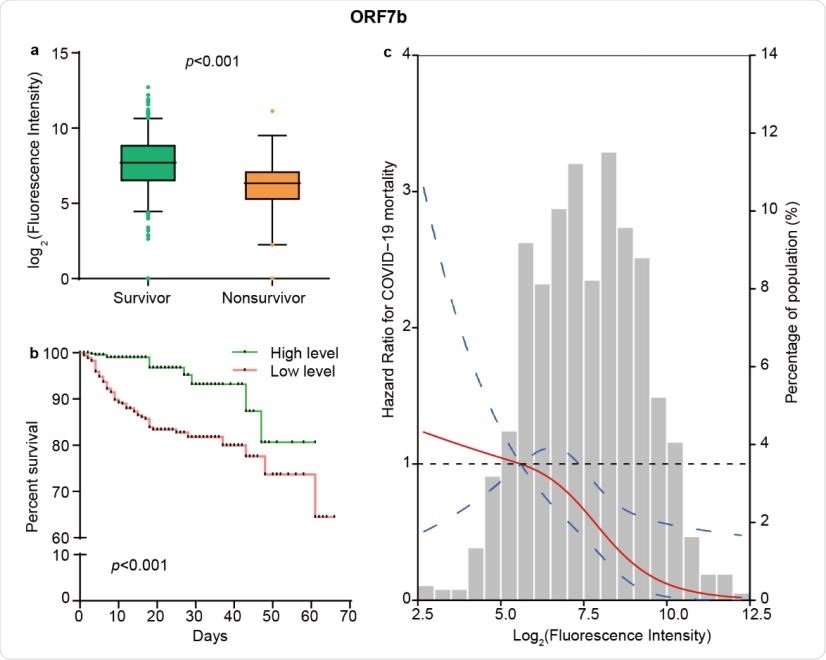
The levels of ORF7b IgM responses independently predict survival of COVID-19. (a) Comparison of the levels of IgM response to ORF7b between 955 survivors and 79 nonsurvivors. The boxplots show medians (middle line) and the third and the first quartiles (boxes), while the tentacles show 97.5 and 2.5 percentiles of the upper and lower parts of the box. (b) Kaplan-Meier survival curves of patients with different levels of IgM antibody against ORF7b. Based on the median level of ORF7b specific IgM responses, patients were classified as both high and low level groups. (c) The restricted cubic spline for the association between ORF7b IgM and risk of COVID-19 mortality. The lines represent adjusted hazard ratios (HRs) based on restricted cubic splines for the levels of ORF7b IgM in Cox regression model. Knots were placed at the 5th, 50th, and 95th percentiles of the distribution of ORF7b specific IgM levels, and the reference value was set at the 10th percentile. Age, sex, diabetes, hypertension, lymphopenia, increased alanine aminotransferase, and increased lactate dehydrogenase were used as adjustment factors.
IgM / IgG response levels to SARS-CoV-2 proteins are important predictors of patient survival and mortality
The team found that high level of IgM against ORF7b at the time of hospitalization is an independent predictor of patient survival, while IgG response levels to 6 non-structural proteins - NSP4, NSP7, NSP9, NSP10, RdRp (NSP12), NSP14 - and 1 accessory protein - ORF3b - is a significant predictor of patient mortality.
“Our results demonstrate that high level of IgM antibody against ORF7b at the time of hospitalization is an independent predictor of patient survival, while IgG responses to NSP9 and NSP10 possess significant predictive power for patient death.”
These results were accurate even after adjustments for comorbidities, demographics, and common lab markers for disease severity. Spline regression analysis showed that the correlation between NSP9 IgG, ORF7b IgM, and NSP10 IgG and COVID-19 mortality risk is linear. Their areas under curve for predictions were determined using computational cross-validations and were 0.74, 0.66, and 0.68, respectively.
Further validations conducted in the serial samples of these cases showed high accuracy of prediction for clinical outcome. The authors believe that these findings have significant implications for improving clinical management and developing therapeutic interventions and vaccines.
“Our research might improve clinical management and guide the development of effective medical interventions and vaccines by enhancing the further understanding of the pathogenesis of COVID-19.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
SARS-CoV-2 antibody signatures for predicting the outcome of COVID-19 Qing Lei, Cai-zheng Yu, Yang Li, Hong-yan Hou, Zhao-wei Xu, Zong-jie Yao, Yan-di Zhang, Dan-yun Lai, Jo-Lewis Banga Ndzouboukou, Bo Zhang, Hong Chen, Zhu-qing Ouyang, Jun-biao Xue, Xiao-song Lin, Yun-xiao Zheng, Xue-ning Wang, He-wei Jiang, Hai-nan Zhang, Huan Qi, Shu-juan Guo, Mei-an He, Zi-yong Sun, Feng Wang, Sheng-ce Tao, Xiong-lin Fan medRxiv 2020.11.10.20228890; doi: https://doi.org/10.1101/2020.11.10.20228890, https://www.medrxiv.org/content/10.1101/2020.11.10.20228890v2
- Peer reviewed and published scientific report.
Lei, Qing, Cai-zheng Yu, Yang Li, Hong-yan Hou, Zhao-wei Xu, Zong-jie Yao, Yan-di Zhang, et al. 2022. “Anti-SARS-CoV-2 IgG Responses Are Powerful Predicting Signatures for the Outcome of COVID-19 Patients.” Journal of Advanced Research 36 (February): 133–45. https://doi.org/10.1016/j.jare.2021.11.014. https://www.sciencedirect.com/science/article/pii/S2090123221002332.