The coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pathogen, continues to spread across the globe.
As previous studies have shown, over the course of the pandemic, SARS-CoV-2 has evolved and mutated several times.
A team of researchers from the School of Information Sciences in Illinois, USA, has revealed the rate of mutation in the 29 proteins that make up the SARS-CoV-2 microbe.
The researchers found that SARS-CoV-2's spike protein mutation has slowed down over the several months of the pandemic, offering hope to the promising vaccine candidates nearing the end of their clinical trials.
On the other hand, there are multiple regions of intrinsic disorder on the virus's protein structure, which could become a problem for the long-term efficacy of these vaccines.
Study background
In December 2019, the first case of COVID-19 was identified in Wuhan City in China. Since then, the novel virus has spread to 191 countries and territories. With over 58.7 million confirmed cases, over 1.38 million people have died due to the virus.
SARS-CoV-2 is a single-stranded RNA-enveloped virus. A characterizing feature of SARS-CoV-2 is the presence of protein spikes that cover its surface, which the virus uses to bind with and enter human cells.
Called spike proteins (or s proteins), these spikes bind with the angiotensin-converting enzyme 2 (ACE2) receptors found abundantly in human epithelial cells. The ACE2 acts as a cellular gateway for the virus to enter and cause infection.
The SARS-CoV-2, just like other viruses, mutates as it spreads across the globe. Viral genome sequence data has been collected in real-time from COVID-19 patients at a significant pace. By May, the GISAID has gathered more than 15,000 full sequences to study when, where, and how mutations happen within the virus's genome.
To date, most gene studies focus on the S-protein, which has revealed that the receptor-binding domain (RBD) is the most variable region, with many RBD amino acids showing crucial ACE2 receptor binding functions. However, the mutations that occur in other genomic regions have been rarely tackled.
Coronaviruses (CoVs) contains four structural proteins, including the Spike (S), Membrane (M), Envelope (E), and Nucleocapsid (N). The spike protein, a trimeric glycoprotein of coronaviruses, mediates the coronaviruses' binding to the host cell's surface-specific receptors.
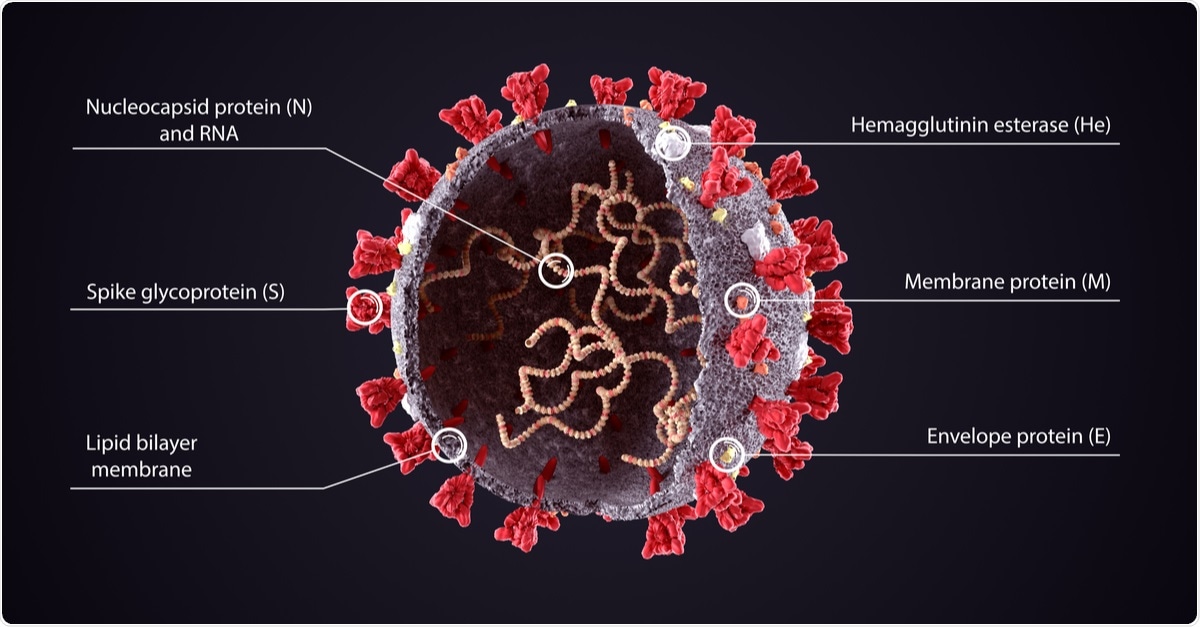
3D Illustration of COVID-19 Virus Structure Diagram. Corona Virus SARS-CoV-2, 2019 nCoV virus sheme. Full text description with sliced model and RNA on dark background. Image Credit: Orpheus FX / Shutterstock
The N-protein is the most abundant in both viruses and virus-infected cells. It plays many roles in the replication and transcription of the virus, including the assembly of the viral terminal end. It helps form the ribonucleoprotein complex that works in maintaining a functional RNA conformation. Mutations in this protein could alter both virulence and transmissibility
The study
The study, which appeared in the journal Evolutionary Bioinformatics, highlighted the genomic accumulation of mutations at various points early on in the pandemic. This way, the researchers could identify changes in mutationally highly active genomic regions across the globe.
To arrive at the study’s findings, the researchers used the Wuhan NC-045512.2 sequence as a reference. From there, they sampled 15,342 indexed sequences from GISAID, translating them into proteins and grouping them by the month of deposition.
The researchers described new pathways of entropic expansion of mutations that involve intrinsically disordered regions of the SARS-CoV-2 proteome. Pathways involve intrinsically disordered regions of the N-protein, which interacts with the M-protein during viral assembly. When this happens, the virus’s transcription efficiency is enhanced, helping it to overcome the host’s innate immune response.
Further, the researchers discovered dominant variants, which are found on the protein surface. Mutation entropy also decreased from March to April, after surges at various sites, including the D614G mutation site of the S-protein.
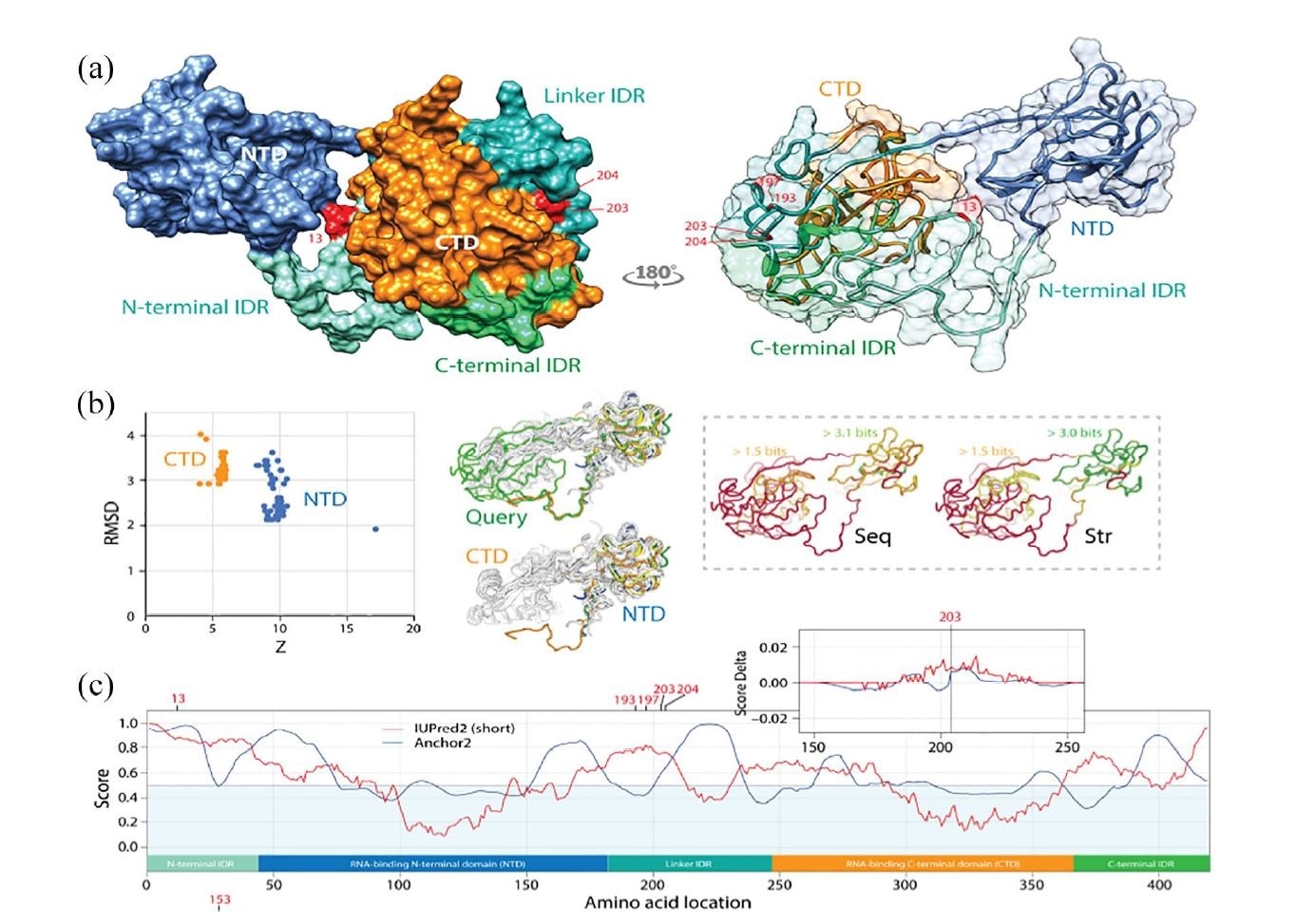
Pathways of mutational diversification of SARS-CoV-2 involve intrinsic disordered regions of the nucleocapsid (N) protein. (a) The N-protein has 2 major RNA-binding domains, an N-terminal domain (NTD) and a C-terminal domain (CTD), both connected to a central linker and flanked by terminal sequences, all of which have been reported to be intrinsically disordered regions (IDRs). Mutations were traced onto a SARS-CoV-2 N-protein structure modeled with I-Tasser. They occurred in position 13 of the N-terminal IDR and positions 193, 197, 203 and 204 of the linker IDR, all of them in loop regions of the molecule. Mutations 203 and 204 were the only sites that were buried in the molecule. (b) A DALI structural neighborhood analysis against the modeled structure (88 structural neighbors, including many from SARS-CoV-2) showed 2 clusters in the RMSD versus Z-score plot, one reflecting structural match to the NTD domain and the other to the CTD domain. Structural alignment plots of the 88 structures supported the veracity of the modeled RNA-binding domains and revealed that the NTD is more conserved at sequence (Seq) and structure (Str) levels. (c) The mapping of intrinsic disorder (UIPred2, red line) and gain-loss of binding energy (Anchor2, blue line) along the sequence confirmed the significant intrinsic disorder and binding (scores ? 0.5) of linker and terminal regions. A comparison of the R203K mutant and reference viral strain with a delta score revealed that the mutation increased disorder. A similar outcome was obtained with the G204R mutant.
The team also noted expanding mutations of the R203L and G204R of the N-protein’s inter-domain region between March and April. The regions with these mutations showed marked intrinsic disorder, which was enhanced by the N-protein.
"The study provides valuable information for therapeutics and vaccine design, as well as insight into mutation tendencies that could facilitate preventive control," the researchers noted in the study.
Conclusion
The good news is, the study findings show that potential vaccines, which are undergoing clinical trials or regulatory body approval, may offer hope in combatting COVID-19. However, the team also found that multiple regions of intrinsic disorder on other proteins may cause future problems for these vaccines.
Future studies and vaccine developments may benefit from these fundings.
The researchers have said that the COVID-19 genetic tracking is not about eradicating the virus but staying on top of the changes long enough for the body's immune system to break it down into something more manageable and less virulent.
"Some coronaviruses live with us and do not cause disease. That's the best option for the virus. It wants to strike a balance between very aggressive and very mild. This thing came from animals, so it is an oddity and is trying to reach that equilibrium," Gustavo Caetano-Anolles, an author of the study, explained.
He added that studying genetic mutations of the novel virus and exploring what happened in the first wave can help in the battle against the surging second and third waves of the infection.