Since effective antiviral treatment and fully validated COVID-19 vaccines are thus far unavailable in our battle against COVID-19, precise and accurate quantification of the SARS-CoV-2 virus plays a pivotal role in appraising infectivity parameters and controlling the ongoing pandemic.
In order to quantify SARS-CoV-2, probe-based reverse transcription-polymerase chain reaction (RT-PCR) is currently used as the gold standard technique due to its relatively high sensitivity and specificity; nonetheless, it depends on expensive instruments and well-designed probes not amenable for small clinics or community health environments.
Likewise, isothermal amplification methods are actually intended for qualitative detection purposes or frequently exposed to either undesired non-specific amplification or false positive signals.
This is where CRISPR-Cas-based nucleic acid detection comes into play. Still, the question is whether it can be tinkered to enable accurate quantification without undesired premature target amplification and subsequent overestimation if digital detection is used.
A research group from the United States, led by Dr. Xiong Ding from the Department of Biomedical Engineering at the University of Connecticut Health Center in Farmington, described a digital WS-CRISPR assay that promises valid and reliable quantification of SARS-CoV-2 nucleic acids in clinical samples.
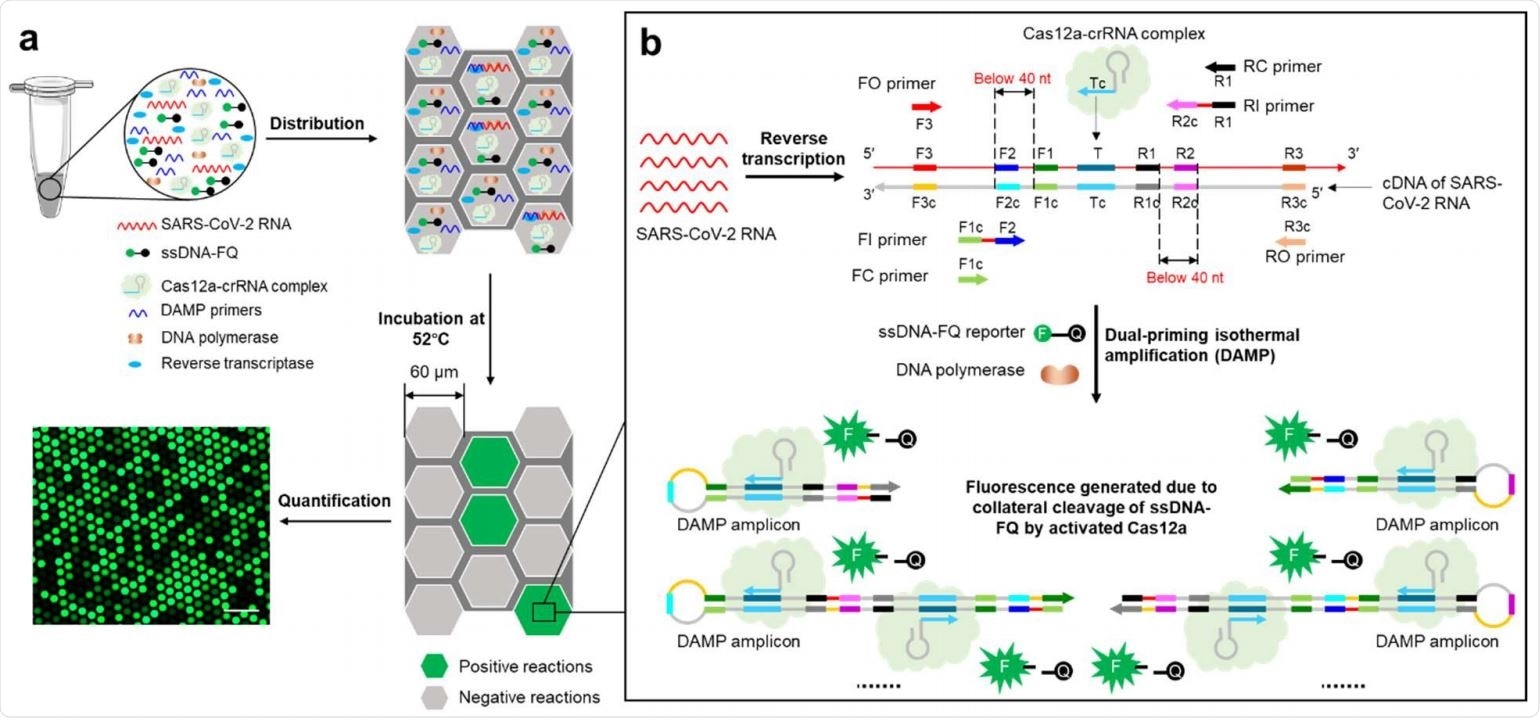
Overview of digital WS-CRISPR assay. a, One-pot WS-CRISPR reaction mixture is first prepared in one tube. After distributed into QuantStudio 3D digital chip, over ten thousand sub-nanoliter (~0.7 nL) microreactions are isolated in microwells. When incubated at 52°C, each microreaction with SARS-CoV-2 RNA target undertakes WS-CRISPR reaction and generates strong green fluorescence (positive spots), whereas not in those without target (negative spots). Scale bar is 300 μm. Through detecting and counting the positive microreactions (or spots), SARS-CoV-2 RNA can be quantified based on the proportion of positive spots. b, Working principle of one-pot WS-CRISPR assay for SARS-CoV-2 detection. The WSCRISPR reaction mixture contains Cas12a-crRNA complex, six DAMP primers (two outer primers of FO and RO, two inner primers of FI and RI, and two competition primers of FC and RC), ssDNA-FQ reporter, SuperScript IV reverse transcriptase, Bst DNA polymerase, and SARS-CoV-2 RNA in a one-pot format.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Building blocks of the new method
In this exciting scientific endeavor, the researchers took advantage of an inaugural one-pot warm start CRISPR reaction, which combines a low-temperature reverse transcription dual-priming mediated isothermal amplification (RT-DAMP) and CRISPR-Cas12a-based detection.
More specifically, to couple these two different reaction systems into one-pot, pyrophosphatase has been added to maintain a constant concentration of magnesium ions by degrading the magnesium pyrophosphate byproduct.
In addition, phosphorothioated inner primers have been used to enable efficient reverse transcription isothermal amplification at rather low-temperatures (such as 52 °C) in this study.
Finally, by partitioning the one-pot reaction mix into sub-nanoliter microreactions using QuantStudio 3D digital chips, this research group developed a digital CRISPR assay that enables sensitive and dependable quantification of SARS-CoV-2.
Moving the goalpost of CRISPR diagnostics
In comparison to previously reported CRISPR-based nucleic acid techniques, this digital WS-CRISPR assay provides several exceptional advantages. First and foremost, this is an inaugural demonstration of a one-pot CRISPR assay that combines Bacillus stearothermophilus (Bst) DNA polymerase-based reverse transcription isothermal amplification with CRISPR-Cas12a detection – without the need for higher reaction temperature and longer nucleic acids.
Furthermore, the method is typically initiated at an elevated temperature (above 50 °C), addressing the issue of undesired premature target amplification in digital detection, as well as displaying 10-fold higher sensitivity and high detection specificity when compared to tube-based bulk assay format.
By targeting the nucleoprotein gene of SARS-CoV-2, it is possible to quantify as low as 5 copies of SARS-CoV-2 RNA per microliter in the chip with the use of digital WS-CRISPR assay. Moreover, the quantification can occur in clinical samples, enabling the assessment of COVID-19 infectivity and antiviral drug efficacy.
Finally, the proposed digital WS-CRISPR assay displays a high tolerance level to inhibitors and can be used to directly detect the virus in crude saliva samples, circumventing the need for RNA extraction. This will, in turn, facilitate the COVID-19 diagnosis and lower the infection risk in health workers.
A glimpse into the future
"As the first clinically validated digital CRISPR assay, our digital WS-CRISPR assay provides a reliable, sensitive and straightforward SARS-CoV-2 quantitative detection", emphasize study researchers in this medRxiv preprint paper. "It opens a new exploration for CRISPR-base nucleic acid quantitative detection," they add.
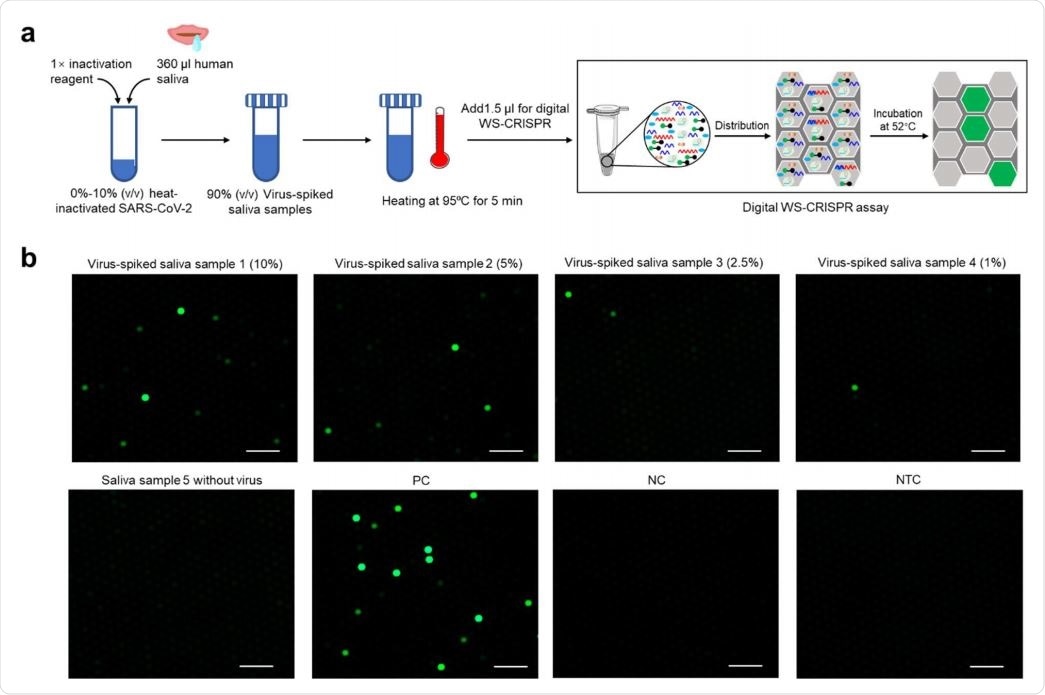
Direct detection of SARS-CoV-2 in crude saliva samples by digital WS-CRISPR assay. a, Workflow for direct SARS-CoV-2 testing in spiked saliva samples by digital WS-CRISPR assay. b, Endpoint fluorescence micrographs of the chip for direct detection of SARS-CoV-2 virus spiked in saliva samples. Saliva samples 1-5, the samples with 10%, 5%, 2.5%, 1%, and 0% of heat-inactivated SARS-CoV-2 virus. Each micrograph is a representative of six distinct regions taken to cover about 2809 microreactions. PC, SARS-CoV-2-positive control sample. NC, SARS-CoV-2-negative control sample. NTC, non-template control. Scale bars are 300 μm.
Notwithstanding the advantages above, a new digital chip has to be further explored and validated for digital WS-CRISPR assay in the future, with the need for additional clinical validation steps.
In addition, albeit this digital WS-CRISPR assay utilizes relatively expensive fluorescence microscopy for imaging purposes, smartphone-based portable fluorescence microscopy may become the alternative option towards onsite quantitative detection in the near future.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources