Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – the causative pathogen of coronavirus disease 2019 (COVID-19) – has infected over 61.1 million people worldwide and caused over 1.43 million deaths.
As scientists continue to probe for effective pharmaceutical interventions against SARS-CoV-2, a new study reports a very promising monoclonal antibody, HB27, that appears to have two distinct mechanisms of action inhibiting the virus-host cell interaction and subsequent infection. This could therefore be a "promising candidate for immuno-therapies against COVID-19," says the research team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The virus gains entry to host cells via its spike protein, a two-subunit protein that engages the angiotensin-converting enzyme 2 (ACE2) receptor on the host cell membrane. This exposes the protein to host cell proteases, which then triggers a massive change in conformation from pre-fusion to post-fusion stages. This leads to the fusion of the viral and host cell membranes to allow the virus to enter the cytosol of the host cell.
The S protein exists as a trimer in nature, but has a receptor-accessible and -inaccessible state, wherein the RBDs are open (one or more) or closed (all). These states are possible because of the hinge-like movement at the RBD mediated by complex protein-protein interactions with host cells.
Though neutralizing antibodies (NAb) have been identified, some, like CR3022, fail to prevent infection with the virus. This is probably due to the flexible conformation of the neutralizing epitopes, which enable the infection to proceed despite the presence of NAbs. On the other hand, other antibodies are being discovered which appear to neutralize the virus without blocking the ACE2 receptor.
The current study, published in the bioRxiv* server in November 2020, aims at understanding the mechanism of neutralization of SARS-CoV-2 by a newly identified monoclonal antibody. The NAb was identified from an antibody library obtained from mice immunized with recombinant SARS-CoV-2 RBD, and was a chimeric one, named MHB27. Since this had powerful binding and neutralizing activity against the virus, a humanized form was generated, called HB27.
Both the IgG and HB27 forms of this antibody bind to the RBD of SARS-CoV-2 at low and very low nanomolar affinity, but not to the RBD of the earlier SARS-CoV and MERS-CoV. Its half-maximal inhibitory concentration (IC50) is also in the very low nanomolar range. It was further tested against the wildtype virus in a plaque reduction neutralization test (PRNT), and showed a nanomolar neutralizing concentration.
Mouse infection prevented
In susceptible mice, a single dose of HB27 was able to prevent infection with the mouse-adapted SARS-CoV-2, MASCp6, when given either before or after exposure to the virus, with the viral load being reduced by over 99.9% in lungs and trachea.
They then tested the ability of HB27 to prevent SARS-CoV-2 infection in a mouse model adapted to express human ACE2 receptors. They found, again, that this molecule was able to reduce viral loads in the lungs and trachea at 5 days following challenge with the virus, if the antibody was administered either before or after exposure. However, preventive dosing was more powerful, with the lung viral levels being reduced over a thousand-fold.
The low levels of viral RNA seen in the lungs and trachea at days 3 and 5 post-infection may be because of the intranasal challenge, where IgG antibodies cannot bind to the virus directly, allowing it to enter the lungs and trachea. However, no infectious virions could be found in these tissues at these time points, as shown by a PRNT.
While the control mice showed signs of moderate interstitial pneumonia, the HB27-treated mice had minimal or very mild inflammatory infiltrates, showing the ability of the NAb to prevent or treat the infection following exposure to SARS-CoV-2.
Non-toxic at high doses
The researchers also tested the antibody's toxicity by giving it at a single high dose, intravenously, at 10 times the estimated effective dose in humans, to the non-human primate (NHP) model, rhesus macaques. Of the four tested animals, none showed any signs of toxicity, and the half-life was found to be about 10 days, on average. Interestingly, this period is probably adequate for its therapeutic efficacy, since at 5 days, there was a 99.9% reduction in viral load. The results indicated its safety in NHPs.
"Double Lock" on SARS-CoV-2
The scientists then investigated the mechanism of neutralization, and found that HB27 completely prevented soluble ACE2-trimeric S interaction. Not only so, already bound soluble ACE2 was replaced by HB27, because the latter has a thousand times stronger binding affinity to the spike protein. However, much higher concentrations are required to neutralize this binding when the ACE2 receptor is on the cell surface, perhaps because it is dimeric. When tested by real-time polymerase chain reaction reverse transcription (RT-PCR), the researchers confirmed that cells treated with HB27 before and after virus attachment at below-micromolar concentrations could displace already bound virions at a concentration of about 2.5 nM.
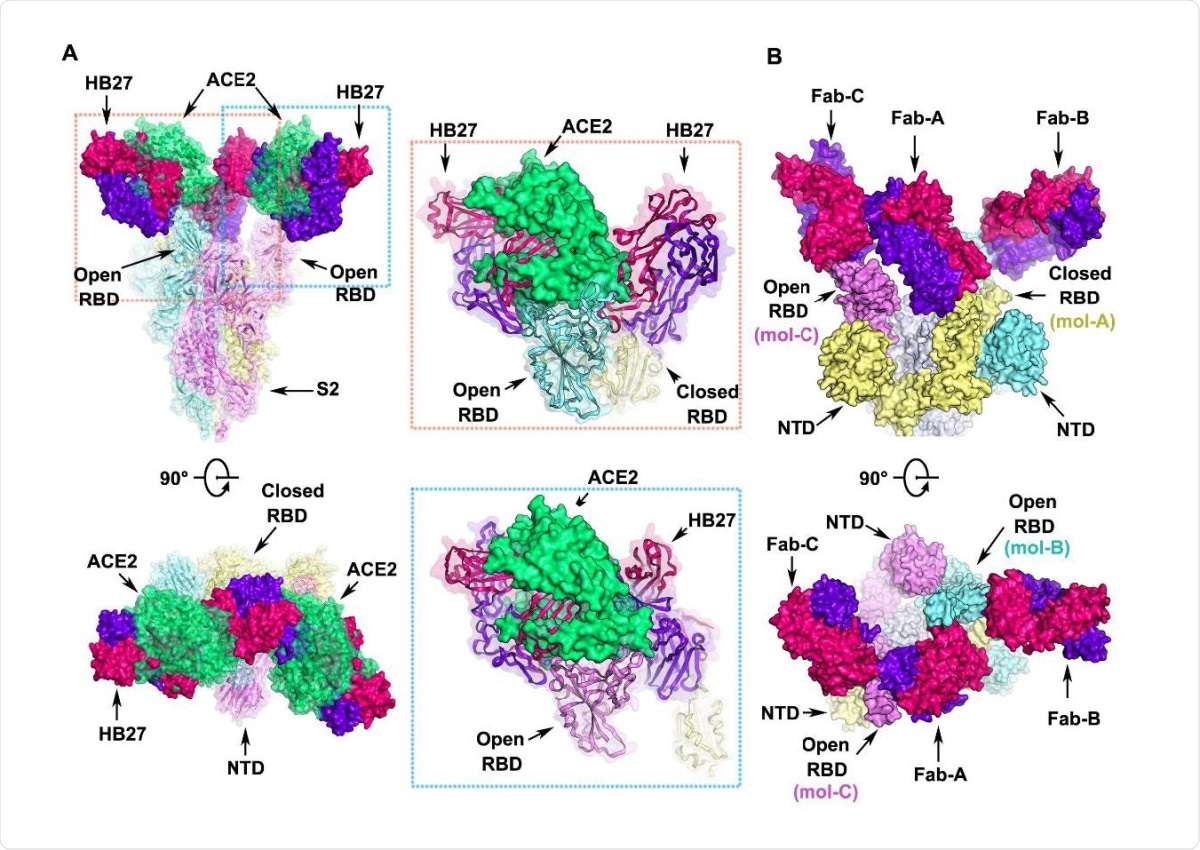
Structural basis for neutralization of SARS-CoV-2 by HB27 (A) Orthogonal views of the clashes between HB27 Fabs and ACE2 upon binding to SARS-CoV-2 S trimer. The SARS-CoV-2 S trimer is presented as ribbon diagrams and translucent molecular surfaces with three monomers colored in cyan, yellow and violet, respectively. The three copies of HB27 Fabs are rendered as molecular surfaces colored 636 the same as in Figure 6. The superposed ACE2 is presented as green ribbon diagrams as well as translucent molecular surface. Insets are close-up views of the clashes between ACE2 and HB27 upon binding to SARS-CoV-2 RBD. (B) Orthogonal views of the structure of HB27 Fab-A, Fab-B and Fab-C complexed with SARS-CoV-2 RBD. The S1 subunits of SARS-CoV-2 S trimer are rendered as cyan, yellow and violet surfaces and the S2 subunits are rendered as gray surfaces.
Secondly, they found that HB27 also inhibits fusion of viral and host cell membranes. They used a spike-mediated cell-cell fusion system with a cell line that expresses SARS-CoV-2 and a GFP tag as the effector cells that propagate infection, and Vero E6 cells as the target cells that suffer infection. They found that when both cell types were incubated, hundreds of cells fused to form a large syncytial cell. However, the presence of HB27 prevented this completely, at a concentration of 0.5 μM. This was not seen with another neutralizing antibody, H104.
They also carried out neutralization assays, conducted by introducing HB27 after the virus had already bound to its receptor. They found that it prevented or reduced syncytium formation, depending on the dose at which it was added, from 4-100 nM, and at 100 nM, complete inhibition was observed. It is also capable of blocking the pH-dependent fusion of the virus with liposome membranes in a dose-dependent manner, which offers another mechanism of neutralization, though at a higher concentration.
The researchers conclude that the main mechanism of neutralization is likely to be inhibiting virus-receptor attachment. The inhibition of membrane fusion may depend on whether the molecule can gain access to the endosome containing the virus.
Mechanism of inhibition
They then did a structural visualization of the inhibition process. The RBD head is inserted into a cavity formed by five CDR regions, via many hydrophobic interactions, with five additional hydrogen bonds. The antigenic site to which HB27 binds has 12 residues, only 7 being common to SARS-CoV-2 and SARS-CoV, which may explain why the antibody cannot neutralize the latter. Again, of the RBD mutations reported so far, none lie within this epitope, and all RBD mutants had similar binding affinities on testing with HB27, as did the D614G mutant.
The neutralization mechanism is by steric hindrance of ACE2 binding, with two adjacent HB27 molecules sterically preventing the binding of one ACE2 molecule. The trimeric S was observed to be in only one conformational state, of one closed with two open. The Fab of the HB27 attached to the closed RBD appears to anchor all three RBDs and keeps them from changing.
Unlike several other cross-reactive monoclonal antibodies with different patterns of neutralization, involving conserved patches of amino acids near the edge of the RBM but not within it, HB27 is at the more variable edge of the RBM core and occupies all RBDs. This could explain why this antibody is specific and highly neutralizing.
Conclusion
The researchers conclude, "HB27 probably inhibits SARS-CoV-2 infection at multiple steps during the viral entry process." This is the first antibody reported to be capable of preventing membrane fusion by a coronavirus. Moreover, in line with some recent research, the disruption of host cell-virus attachment by this antibody could prevent the association between the virus and the host cell protease TMPRSS2; again, possibly, preventing viral membrane fusion by inhibiting TMPRSS2-mediated spike cleavage.
The authors describe this: "The potent neutralizing activity of HB27 probably results from its intervention at two steps of viral infection, locking away attachment of the virus to its receptor and blocking membrane fusion; resulting in a double lock." The results of the ongoing clinical trials of this antibody in China should be full of interest.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources