An interesting new study throws a flood of light on how the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus accomplishes its mission to infect and replicate within host cells while evading the host immune response. The study, published as a preprint on the bioRxiv* server in November 2020, shows how the virus not only suppresses translation of cellular proteins but increases the rate of breakdown of cellular messenger ribonucleic acids (mRNAs) that defend the host against the virus.
Novel Coronavirus SARS-CoV-2 Colorized scanning electron micrograph of an apoptotic cell (blue) infected with SARS-COV-2 virus particles (red), isolated from a patient sample. Image captured at the NIAID Integrated Research Facility (IRF) in Fort Detrick, Maryland. Image Credit: NIAID / Flickr
The virus has a single-stranded ribonucleic acid (RNA) genome of about 30 kb, which encodes two polyproteins on two open reading frames, ORF1ab. These are translated into peptides that are cleaved into 16 functional viral proteins eventually. These are the nonstructural proteins (nsps) like RNA-dependent RNA polymerase (RdRp), which mediates the transcription of subgenomic RNA into the same 5’ leader sequence but different segments of the 3’ end of the viral RNA genome.
These different segments of subgenomic RNA encode the structural viral proteins such as the spike (S) and nucleocapsid (N) proteins, as well as accessory proteins. Such proteins can be translated from the subgenomic RNA only by the cell’s own translation apparatus, which is hijacked by the virus for its own purposes. The virus simultaneously keeps the host immune response at bay.
Firstly, the infected cell uses the interferon (IFN) pathway. The infected cell senses the presence of viral RNA transcripts and responds by promoting the entry of transcription factors into the nucleus to induce the production of type I IFNs. These bind to their receptors on the same and other cells to increase the signal strength, thus causing the secretion of hundreds of IFN-stimulated genes (ISGs) that hinder viral replication at different points.
The IFNs are thought to be very important in the disease process following SARS-CoV-2 infection. The current study aimed at producing a picture of how the virus interferes with host cell translation, using RNA sequencing and ribosomal profiling to monitor both RNA production (transcription) and protein synthesis (translation) simultaneously. The experiment was carried out in duplicate to ensure the results were reproducible.
Global inhibition of protein translation
SARS-CoV-2 is unique in the degree of inhibition of IFN signaling. This occurs by shutting off host protein synthesis, mainly by the viral nsp1. This prevents translation by binding to the 40S ribosome, closing off the mRNA entry channel of the ribosomes. Since fewer ribosomes are available for translation, there is a global reduction in protein synthesis.
At 3 hours following infection, viral mRNA was translated at the same kind of efficiency as the host mRNA (similar translation efficiency, TE). At 8 hours post-infection, TE was shifted significantly to inhibit viral transcripts. This reduced translation efficiency perhaps because some viral mRNAs are not available for translation, being enclosed in the double-membrane compartments within which replication occurs.
This reduction in TE was focused on the ORFs at the 3’ end but not those in the middle, while those at the 5’ end had an increase in their TE relative to the others. The 5’ UTR being the same for all these ORFs, this may be because of either differing length of viral transcripts or some function of the 3’ UTR.
Using an indicator to measure nascent protein synthesis levels, the researchers found that overall, translation was already reduced at 3 hours post-infection, declining further over time until it was only 50% normal. However, total RNA remained unchanged, as did ribosomal RNA (rRNA).
By 8 hours post-infection, the amount of cellular and viral mRNA (indicating transcription) exceeded that of rRNA (indicating translation), due to high viral transcription. At the same time, the cellular mRNA was reduced to half. Some cellular mRNAs, however, were at higher levels, including those related to the immune response like TLR signaling, chemokines and cytokines such as IL6 and IL8, as well as several ISGs.
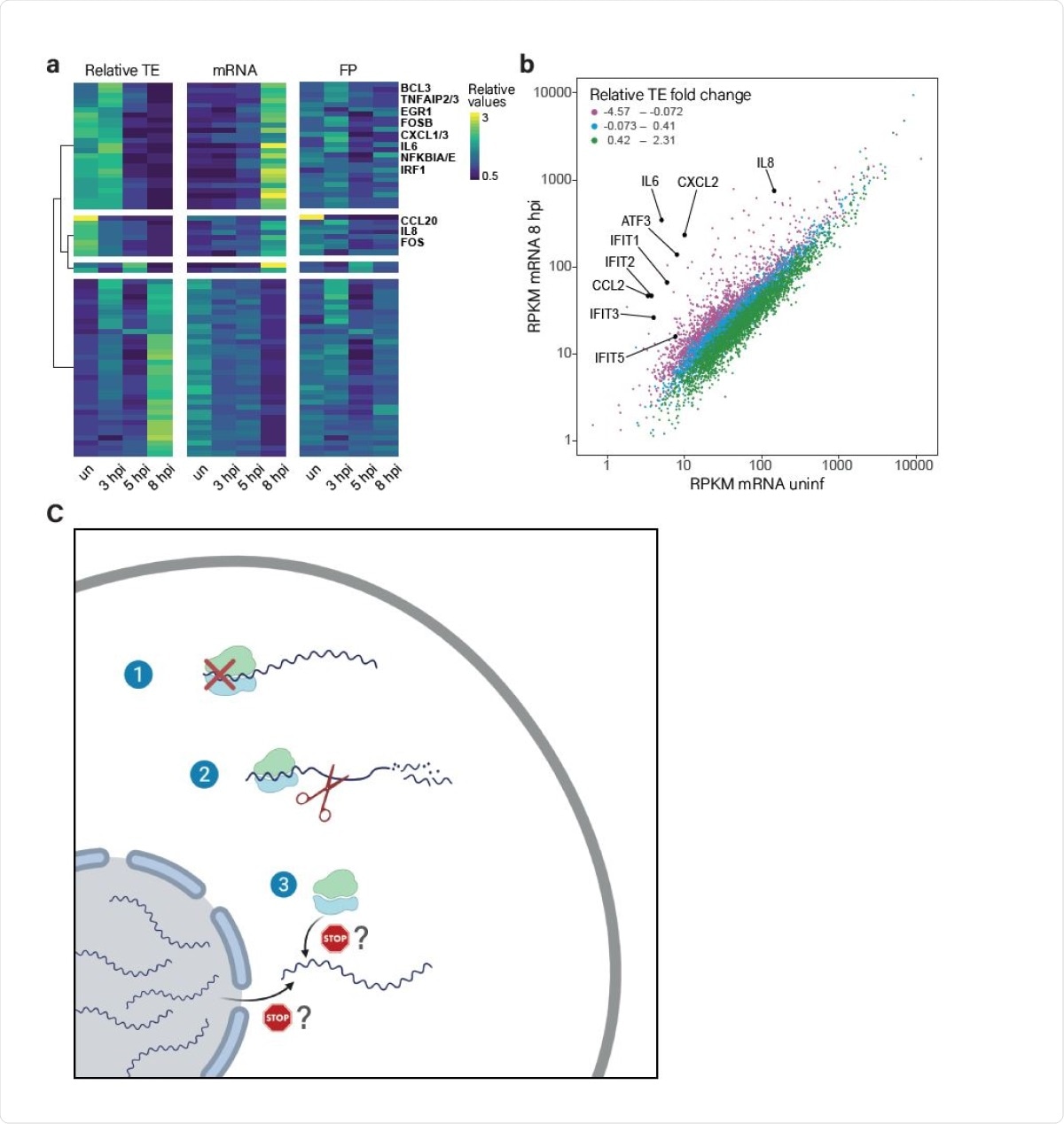
The translation of induced transcripts is impaired during infection. (A) Heat map presenting relative TE, mRNA and footprints (FP) of human genes that showed the most significant changes in their relative TE along SARS-CoV-2 infection. Shown are relative expression ratios after partitioning clustering based on changes in relative TE values. (B) Scatter plot presenting cellular transcript levels in uninfected cells compared to 8hpi. Genes are colored based on the relative change in their TE between uninfected and 8hpi. Central cytokines and IFN stimulated genes are labeled. (C) A model of how SARS-CoV-2 suppresses host gene expression through multi-pronged approach: 1. Global translation reduction; 2. Degradation of cytosolic cellular mRNAs; 3. Specific translation inhibition of newly synthesized cellular mRNAs.
Thus, host protein synthesis was shut off by a “general reduction in the translation capacity of infected cells and reduction in the levels of most cellular mRNAs.”
Degradation of cytosolic cellular transcripts
The cellular transcripts that were reduced were mostly cytosolic, rather than nuclear, especially those being translated. Mitochondrial transcripts were also relatively spared. The scientists observed that the viral nsp1 not only shuts off host cell translation on a broad scale, but accelerates the breakdown of cellular mRNAs. They also found that this increased turnover of mature cellular transcripts causes exonic reads to be reduced at a higher rate relative to intronic reads.
Viral RNA transcribed from its genome resists this breakdown, probably protected by their 5’ UTR regions. This finding is important, in that previous studies showed that the viral 5’ UTR helped viral mRNAs to be translated more efficiently by evading the global inhibition of translation. The current experiments showed that the increase in viral mRNAs was not due to the preferential translation of viral RNA in infected cells, but rather the 5’-UTR-mediated refractoriness of these mRNAs to nsp1-induced degradation.
The total mRNA content was made up of approximately 90% viral RNA, and only 10% cellular mRNA. However, at the ribosomal level, only a third of the RNA bound to these organelles (indicating active translation to proteins) comprised viral mRNA. The effect is that viral translation products become dominant in the mRNA pool within the cell.
Specific translation inhibition of newly transcribed mRNAs
Finally, the virus preferentially inhibits the translation of genes that are induced by the infection, including those concerned with innate immunity. Therefore, despite the increase in RNA, its inability to achieve an increase in translation may explain how the IFN response fails.
The researchers concluded that the virus exerts “unprecedented dominance” over the type of transcription and translation that occurs following infection of a host cell. They say, “Disruption of cellular protein production using these three components may represent a multipronged mechanism that synergistically acts to suppress the host antiviral response.”
Other factors like ORF6, which disrupts the transport of molecules from the nucleus to the cytosol, may also help suppress the antiviral response genes, which, though transcribed at higher levels in the nucleus, cannot induce an efficient antiviral response without being exported to the cytosol.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Finkel, Y. et al. (2020). SARS-CoV-2 utilizes a multipronged strategy to suppress host protein synthesis. bioRxiv preprint. doi: https://doi.org/10.1101/2020.11.25.398578.
https://www.biorxiv.org/content/10.1101/2020.11.25.398578v1
- Peer reviewed and published scientific report.
Finkel, Yaara, Avi Gluck, Aharon Nachshon, Roni Winkler, Tal Fisher, Batsheva Rozman, Orel Mizrahi, et al. 2021. “SARS-CoV-2 Uses a Multipronged Strategy to Impede Host Protein Synthesis.” Nature, May. https://doi.org/10.1038/s41586-021-03610-3. https://www.nature.com/articles/s41586-021-03610-3.